A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors
- PMID: 29259199
- PMCID: PMC5736716
- DOI: 10.1038/s41467-017-02347-w
A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors
Abstract
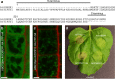
Several Pseudomonas and Xanthomonas species are plant pathogens that infect the model organism Arabidopsis thaliana and important crops such as Brassica. Resistant plants contain the infection by rapid cell death of the infected area through the hypersensitive response (HR). A family of highly related α/β hydrolases is involved in diverse processes in all domains of life. Functional details of their catalytic machinery, however, remained unclear. We report the crystal structures of α/β hydrolases representing two different clades of the family, including the protein SOBER1, which suppresses AvrBsT-incited HR in Arabidopsis. Our results reveal a unique hydrophobic anchor mechanism that defines a previously unknown family of protein deacetylases. Furthermore, this study identifies a lid-loop as general feature for substrate turnover in acyl-protein thioesterases and the described family of deacetylases. Furthermore, we found that SOBER1's biological function is not restricted to Arabidopsis thaliana and not limited to suppress HR induced by AvrBsT.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

