The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy
- PMID: 29259203
- PMCID: PMC5736707
- DOI: 10.1038/s41467-017-02329-y
The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy
Abstract
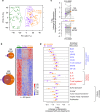
Treatment of chronic lymphocytic leukemia (CLL) has shifted from chemo-immunotherapy to targeted agents. To define the evolutionary dynamics induced by targeted therapy in CLL, we perform serial exome and transcriptome sequencing for 61 ibrutinib-treated CLLs. Here, we report clonal shifts (change >0.1 in clonal cancer cell fraction, Q < 0.1) in 31% of patients during the first year of therapy, associated with adverse outcome. We also observe transcriptional downregulation of pathways mediating energy metabolism, cell cycle, and B cell receptor signaling. Known and previously undescribed mutations in BTK and PLCG2, or uncommonly, other candidate alterations are present in seventeen subjects at the time of progression. Thus, the frequently observed clonal shifts during the early treatment period and its potential association with adverse outcome may reflect greater evolutionary capacity, heralding the emergence of drug-resistant clones.
Conflict of interest statement
C.J.W. is founder of Neon Therapeutics and a member of its scientific advisory board. A.W. and J.A.B. received research funding from Pharmacyclics, an Abbvie Company. J.A.B. received research funding from Gilead. A.W. received research funding from Acerta Pharma. The remaining authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

