A mutant of phosphomannomutase1 retains full enzymatic activity, but is not activated by IMP: Possible implications for the disease PMM2-CDG
- PMID: 29261720
- PMCID: PMC5736207
- DOI: 10.1371/journal.pone.0189629
A mutant of phosphomannomutase1 retains full enzymatic activity, but is not activated by IMP: Possible implications for the disease PMM2-CDG
Abstract
The most frequent disorder of glycosylation, PMM2-CDG, is caused by a deficiency of phosphomannomutase activity. In humans two paralogous enzymes exist, both of them require mannose 1,6-bis-phosphate or glucose 1,6-bis-phosphate as activators, but only phospho-mannomutase1 hydrolyzes bis-phosphate hexoses. Mutations in the gene encoding phosphomannomutase2 are responsible for PMM2-CDG. Although not directly causative of the disease, the role of the paralogous enzyme in the disease should be clarified. Phosphomannomutase1 could have a beneficial effect, contributing to mannose 6-phosphate isomerization, or a detrimental effect, hydrolyzing the bis-phosphate hexose activator. A pivotal role in regulating mannose-1phosphate production and ultimately protein glycosylation might be played by inosine monophosphate that enhances the phosphatase activity of phosphomannomutase1. In this paper we analyzed human phosphomannomutases by conventional enzymatic assays as well as by novel techniques such as 31P-NMR and thermal shift assay. We characterized a triple mutant of phospomannomutase1 that retains mutase and phosphatase activity, but is unable to bind inosine monophosphate.
Conflict of interest statement
Figures


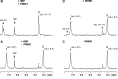


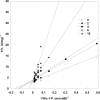


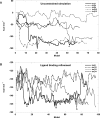
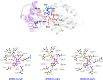
References
-
- Matthijs G, Schollen E, Pardon E, Veiga-Da-Cunha M, Jaeken J, Cassiman JJ, et al. Mutations in PMM2, a phosphomannomutase gene on chromosome 16p13, in carbohydrate-deficient glycoprotein type I syndrome (Jaeken syndrome). Nat Genet. 1997;16(1):88–92. doi: 10.1038/ng0597-88 . - DOI - PubMed
-
- Jaeken J, Hennet T, Freeze HH, Matthijs G. On the nomenclature of congenital disorders of glycosylation (CDG). J Inherit Metab Dis. 2008;31(6):669–72. Epub 2008/10/25. doi: 10.1007/s10545-008-0983-x . - DOI - PubMed
-
- Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833(11):2430–7. Epub 2013/04/16. doi: 10.1016/j.bbamcr.2013.04.001 [pii]. . - DOI - PubMed
-
- Maeda Y, Kinoshita T. Dolichol-phosphate mannose synthase: structure, function and regulation. Biochim Biophys Acta. 2008;1780(6):861–8. Epub 2008/04/05. doi: 10.1016/j.bbagen.2008.03.005 [pii]. . - DOI - PubMed
-
- Pirard M, Collet JF, Matthijs G, Van Schaftingen E. Comparison of PMM1 with the phosphomannomutases expressed in rat liver and in human cells. FEBS Lett. 1997;411(2–3):251–4. . - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

