Distribution of Legionella and bacterial community composition among regionally diverse US cooling towers
- PMID: 29261791
- PMCID: PMC5738086
- DOI: 10.1371/journal.pone.0189937
Distribution of Legionella and bacterial community composition among regionally diverse US cooling towers
Abstract
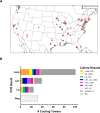
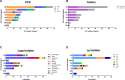
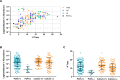
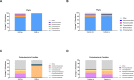
Cooling towers (CTs) are a leading source of outbreaks of Legionnaires' disease (LD), a severe form of pneumonia caused by inhalation of aerosols containing Legionella bacteria. Accordingly, proper maintenance of CTs is vital for the prevention of LD. The aim of this study was to determine the distribution of Legionella in a subset of regionally diverse US CTs and characterize the associated microbial communities. Between July and September of 2016, we obtained aliquots from water samples collected for routine Legionella testing from 196 CTs located in eight of the nine continental US climate regions. After screening for Legionella by PCR, positive samples were cultured and the resulting Legionella isolates were further characterized. Overall, 84% (164) were PCR-positive, including samples from every region studied. Of the PCR-positive samples, Legionella spp were isolated from 47% (78), L. pneumophila was isolated from 32% (53), and L. pneumophila serogroup 1 (Lp1) was isolated from 24% (40). Overall, 144 unique Legionella isolates were identified; 53% (76) of these were Legionella pneumophila. Of the 76 L. pneumophila isolates, 51% (39) were Lp1. Legionella were isolated from CTs in seven of the eight US regions examined. 16S rRNA amplicon sequencing was used to compare the bacterial communities of CT waters with and without detectable Legionella as well as the microbiomes of waters from different climate regions. Interestingly, the microbial communities were homogenous across climate regions. When a subset of seven CTs sampled in April and July were compared, there was no association with changes in corresponding CT microbiomes over time in the samples that became culture-positive for Legionella. Legionella species and Lp1 were detected frequently among the samples examined in this first large-scale study of Legionella in US CTs. Our findings highlight that, under the right conditions, there is the potential for CT-related LD outbreaks to occur throughout the US.
Conflict of interest statement
Figures
References
-
- Mercante JW, Winchell JM. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clinical microbiology reviews. 2015;28(1):95–133. doi: 10.1128/CMR.00029-14 - DOI - PMC - PubMed
-
- Adams D, Fullerton K, Jajosky R, Sharp P, Onweh D, Schley A, et al. Summary of Notifiable Infectious Diseases and Conditions—United States, 2013. MMWR Morbidity and mortality weekly report. 2015;62(53):1–122. doi: 10.15585/mmwr.mm6253a1 - DOI - PubMed
-
- Burillo A, Pedro-Botet ML, Bouza E. Microbiology and Epidemiology of Legionnaire's Disease. Infectious disease clinics of North America. 2017;31(1):7–27. doi: 10.1016/j.idc.2016.10.002 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

