Genome-Based Analysis of Enterococcus faecium Bacteremia Associated with Recurrent and Mixed-Strain Infection
- PMID: 29263205
- PMCID: PMC5824064
- DOI: 10.1128/JCM.01520-17
Genome-Based Analysis of Enterococcus faecium Bacteremia Associated with Recurrent and Mixed-Strain Infection
Abstract
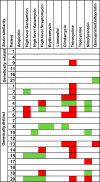
Vancomycin-resistant Enterococcus faecium (VREfm) bloodstream infections are associated with high recurrence rates. This study used genome sequencing to accurately distinguish the frequency of relapse and reinfection in patients with recurrent E. faecium bacteremia and to investigate strain relatedness in patients with apparent VREfm and vancomycin-susceptible E. faecium (VSEfm) mixed infection. A retrospective study was performed at the Cambridge University Hospitals NHS Foundation Trust (CUH) between November 2006 and December 2012. We analyzed the genomes of 44 E. faecium isolates from 21 patients (26 VREfm isolates from 12 patients with recurrent bacteremia and 18 isolates from 9 patients with putative VREfm/VSEfm mixed infection). Phenotypic antibiotic susceptibility was determined using a Vitek2 instrument. Genomes were compared with those of a further 263 E. faecium isolates associated with bacteremia in patients at CUH over the same time period. Pairwise comparison of core genomes indicated that 10 (71%) episodes of recurrent VREfm bacteremia were due to reinfection with a new strain, with reinfection being more likely with increasing time between the two positive cultures. The majority (78%) of patients with a mixed VREfm and VSEfm infection had unrelated strains. More than half (59%) of study isolates were closely related to another isolate associated with bacteremia from CUH. This included 60% of isolates associated with reinfection, indicating acquisition in the hospital. This study provides the first high-resolution insights into recurrence and mixed infection by E. faecium and demonstrates that reinfection with a new strain, often acquired from the hospital, is a driver of recurrence.
Keywords: VRE; mixed infection; recurrence.
Copyright © 2018 Raven et al.
Figures

References
-
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

