Synectin promotes fibrogenesis by regulating PDGFR isoforms through distinct mechanisms
- PMID: 29263300
- PMCID: PMC5752303
- DOI: 10.1172/jci.insight.92821
Synectin promotes fibrogenesis by regulating PDGFR isoforms through distinct mechanisms
Abstract
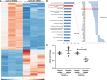
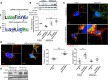
The scaffold protein synectin plays a critical role in the trafficking and regulation of membrane receptor pathways. As platelet-derived growth factor receptor (PDGFR) is essential for hepatic stellate cell (HSC) activation and liver fibrosis, we sought to determine the role of synectin on the PDGFR pathway and development of liver fibrosis. Mice with deletion of synectin from HSC were found to be protected from liver fibrosis. mRNA sequencing revealed that knockdown of synectin in HSC demonstrated reductions in the fibrosis pathway of genes, including PDGFR-β. Chromatin IP assay of the PDGFR-β promoter upon synectin knockdown revealed a pattern of histone marks associated with decreased transcription, dependent on p300 histone acetyltransferase. Synectin knockdown was found to downregulate PDGFR-α protein levels, as well, but through an alternative mechanism: protection from autophagic degradation. Site-directed mutagenesis revealed that ubiquitination of specific PDGFR-α lysine residues was responsible for its autophagic degradation. Furthermore, functional studies showed decreased PDGF-dependent migration and proliferation of HSC after synectin knockdown. Finally, human cirrhotic livers demonstrated increased synectin protein levels. This work provides insight into differential transcriptional and posttranslational mechanisms of synectin regulation of PDGFRs, which are critical to fibrogenesis.
Keywords: Hepatology; Signal transduction.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

