RNA-Seq differential expression analysis: An extended review and a software tool
- PMID: 29267363
- PMCID: PMC5739479
- DOI: 10.1371/journal.pone.0190152
RNA-Seq differential expression analysis: An extended review and a software tool
Abstract
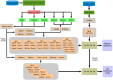
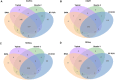
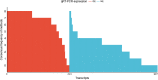
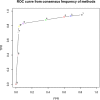
The correct identification of differentially expressed genes (DEGs) between specific conditions is a key in the understanding phenotypic variation. High-throughput transcriptome sequencing (RNA-Seq) has become the main option for these studies. Thus, the number of methods and softwares for differential expression analysis from RNA-Seq data also increased rapidly. However, there is no consensus about the most appropriate pipeline or protocol for identifying differentially expressed genes from RNA-Seq data. This work presents an extended review on the topic that includes the evaluation of six methods of mapping reads, including pseudo-alignment and quasi-mapping and nine methods of differential expression analysis from RNA-Seq data. The adopted methods were evaluated based on real RNA-Seq data, using qRT-PCR data as reference (gold-standard). As part of the results, we developed a software that performs all the analysis presented in this work, which is freely available at https://github.com/costasilvati/consexpression. The results indicated that mapping methods have minimal impact on the final DEGs analysis, considering that adopted data have an annotated reference genome. Regarding the adopted experimental model, the DEGs identification methods that have more consistent results were the limma+voom, NOIseq and DESeq2. Additionally, the consensus among five DEGs identification methods guarantees a list of DEGs with great accuracy, indicating that the combination of different methods can produce more suitable results. The consensus option is also included for use in the available software.
Conflict of interest statement
Figures
References
-
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226 - DOI - PubMed
-
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745 - DOI - PMC - PubMed
-
- Agarwal A, Koppstein D, Rozowsky J, Sboner A, Habegger L, Hillier LW, et al. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC genomics. 2010;11(1):1 doi: 10.1186/1471-2164-11-383 - DOI - PMC - PubMed
-
- Kratz A, Carninci P. The devil in the details of RNA-seq. Nature biotechnology. 2014;32(9):882–884. doi: 10.1038/nbt.3015 - DOI - PubMed
-
- Zhang ZH, Jhaveri DJ, Marshall VM, Bauer DC, Edson J, Narayanan RK, et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PloS one. 2014;9(8):e103207 doi: 10.1371/journal.pone.0103207 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

