Cardiac-specific ablation of the E3 ubiquitin ligase Mdm2 leads to oxidative stress, broad mitochondrial deficiency and early death
- PMID: 29267372
- PMCID: PMC5739440
- DOI: 10.1371/journal.pone.0189861
Cardiac-specific ablation of the E3 ubiquitin ligase Mdm2 leads to oxidative stress, broad mitochondrial deficiency and early death
Abstract
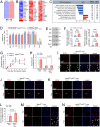
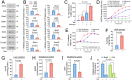
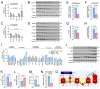
The maintenance of normal heart function requires proper control of protein turnover. The ubiquitin-proteasome system is a principal regulator of protein degradation. Mdm2 is the main E3 ubiquitin ligase for p53 in mitotic cells thereby regulating cellular growth, DNA repair, oxidative stress and apoptosis. However, which of these Mdm2-related activities are preserved in differentiated cardiomyocytes has yet to be determined. We sought to elucidate the role of Mdm2 in the control of normal heart function. We observed markedly reduced Mdm2 mRNA levels accompanied by highly elevated p53 protein expression in the hearts of wild type mice subjected to myocardial infarction or trans-aortic banding. Accordingly, we generated conditional cardiac-specific Mdm2 gene knockout (Mdm2f/f;mcm) mice. In adulthood, Mdm2f/f;mcm mice developed spontaneous cardiac hypertrophy, left ventricular dysfunction with early mortality post-tamoxifen. A decreased polyubiquitination of myocardial p53 was observed, leading to its stabilization and activation, in the absence of acute stress. In addition, transcriptomic analysis of Mdm2-deficient hearts revealed that there is an induction of E2f1 and c-Myc mRNA levels with reduced expression of the Pgc-1a/Ppara/Esrrb/g axis and Pink1. This was associated with a significant degree of cardiomyocyte apoptosis, and an inhibition of redox homeostasis and mitochondrial bioenergetics. All these processes are early, Mdm2-associated events and contribute to the development of pathological hypertrophy. Our genetic and biochemical data support a role for Mdm2 in cardiac growth control through the regulation of p53, the Pgc-1 family of transcriptional coactivators and the pivotal antioxidant Pink1.
Conflict of interest statement
Figures



References
-
- Akazawa H. Mechanisms of cardiovascular homeostasis and pathophysiology—from gene expression, signal transduction to cellular communication. Circ J. 2015;79: 2529–2536. doi: 10.1253/circj.CJ-15-0818 - DOI - PubMed
-
- Borlaug BA, Paulus WJ. Heart failure with preserved ejection fraction: pathophysiology, diagnosis, and treatment. Eur Heart J. 2011;32: 670–679. doi: 10.1093/eurheartj/ehq426 - DOI - PMC - PubMed
-
- Jessup M, Konstam MA, Mancini DM, Michl K, Oates JA, Rahko PS, et al. ACC/AHA 2005 Guideline Update for the Diagnosis and Management of Chronic Heart Failure in the Adult: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Update the 2001 Guidelines for the Evaluation and Management of Heart Failure): developed in collaboration with the American College of Chest Physicians and the International Society for Heart and Lung Transplantation: endorsed by the Heart Rhythm Society. Circulation. 2005;112: e154–e235. doi: 10.1161/CIRCULATIONAHA.105.167586 - DOI - PubMed
-
- Kapiloff MS, Emter CA. The cardiac enigma: current conundrums in heart failure research. F1000Res. 2016;5 doi: 10.12688/f1000research.7278.1 - DOI - PMC - PubMed
-
- Mann DL, Bristow MR. Mechanisms and models in heart failure: The biomechanical model and beyond. Circulation. 2005;111: 2837–2849. doi: 10.1161/CIRCULATIONAHA.104.500546 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

