A coarse-grained model for DNA origami
- PMID: 29267876
- PMCID: PMC5814798
- DOI: 10.1093/nar/gkx1262
A coarse-grained model for DNA origami
Abstract
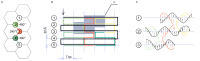
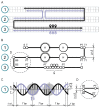
Modeling tools provide a valuable support for DNA origami design. However, current solutions have limited application for conformational analysis of the designs. In this work we present a tool for a thorough study of DNA origami structure and dynamics. The tool is based on a novel coarse-grained model dedicated to geometry optimization and conformational analysis of DNA origami. We explored the ability of the model to predict dynamic behavior, global shapes, and fine details of two single-layer systems designed in hexagonal and square lattices using atomic force microscopy, Förster resonance energy transfer spectroscopy, and all-atom molecular dynamic simulations for validation of the results. We also examined the performance of the model for multilayer systems by simulation of DNA origami with published cryo-electron microscopy and atomic force microscopy structures. A good agreement between the simulated and experimental data makes the model suitable for conformational analysis of DNA origami objects. The tool is available at http://vsb.fbb.msu.ru/cosm as a web-service and as a standalone version.
Figures




References
-
- Ke Y. Designer three-dimensional DNA architectures. Curr. Opin. Struct. Biol. 2014; 27:122–128. - PubMed
-
- Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. - PubMed
-
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964; 5:282–304. - PubMed
-
- Andersen E.S., Dong M., Nielsen M.M., Jahn K., Lind-Thomsen A., Mamdouh W., Gothelf K.V., Besenbacher F., Kjems J.. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano. 2008; 2:1213–1218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

