Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression
- PMID: 29268739
- PMCID: PMC5740586
- DOI: 10.1186/s12931-017-0696-5
Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression
Abstract
Background: Nasal gene expression profiling is a promising method to characterize COPD non-invasively. We aimed to identify a nasal gene expression profile to distinguish COPD patients from healthy controls. We investigated whether this COPD-associated gene expression profile in nasal epithelium is comparable with the profile observed in bronchial epithelium.
Methods: Genome wide gene expression analysis was performed on nasal epithelial brushes of 31 severe COPD patients and 22 controls, all current smokers, using Affymetrix Human Gene 1.0 ST Arrays. We repeated the gene expression analysis on bronchial epithelial brushes in 2 independent cohorts of mild-to-moderate COPD patients and controls.
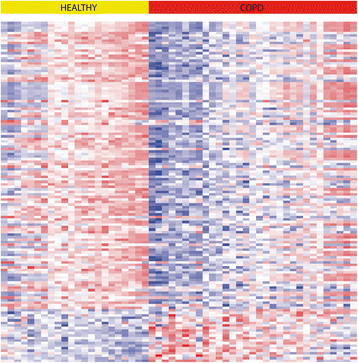
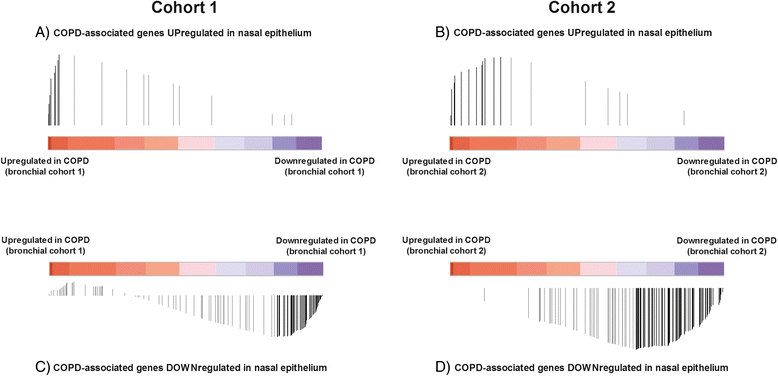
Results: In nasal epithelium, 135 genes were significantly differentially expressed between severe COPD patients and controls, 21 being up- and 114 downregulated in COPD (false discovery rate < 0.01). Gene Set Enrichment Analysis (GSEA) showed significant concordant enrichment of COPD-associated nasal and bronchial gene expression in both independent cohorts (FDRGSEA < 0.001).
Conclusion: We identified a nasal gene expression profile that differentiates severe COPD patients from controls. Of interest, part of the nasal gene expression changes in COPD mimics differentially expressed genes in the bronchus. These findings indicate that nasal gene expression profiling is potentially useful as a non-invasive biomarker in COPD.
Trial registration: ClinicalTrials.gov registration number NCT01351792 (registration date May 10, 2011), ClinicalTrials.gov registration number NCT00848406 (registration date February 19, 2009), ClinicalTrials.gov registration number NCT00807469 (registration date December 11, 2008).
Keywords: Bronchial epithelium; COPD; Genome wide gene expression; Microarray; Nasal epithelium.
Conflict of interest statement
Ethics approval and consent to participate
Identification cohort: The ethical committee of the University Medical Center Groningen (Medisch Ethische Toetsingscommisie) approved the study and all included subjects gave their written informed consent.
Comparator cohort 1: Institutional review board approval was obtained from the participating institutions and all subject gave their written informed consent.
Comparator cohort 2: The ethical committee of the University Medical Center Groningen approved the study and all included subjects gave their written informed consent.
Consent for publication
Not applicable.
Competing interests
MvdB reports research grants paid to the University from Chiesi, GlaxoSmithKline, TEVA and AstraZeneca. HAMK reports fees paid to the University for consultancies/advisory boards and fees per patient for recruitment from GlaxoSmithKline, Boehringer Ingelheim and Novartis. WT reports fees paid to the University from Pfizer, GlaxoSmithKline, Chiesi, Roche Diagnostics/Ventana, Biotest, Merck Sharp Dohme, Novartis, Lilly Oncology and a grant from the Dutch Asthma Fund. PW reports personal fees from GlaxoSmithKline, Boehringer Ingelheim, AstraZeneca and a grant from GlaxoSmithKline. MEL reports grants from NIH/NHLBI during the conduct of the study; in addition, MEL has a patent US PTO 14/406,607 pending. DSP reports consultancy fees or research grants paid to the University from Astra Zeneca, Chiesi, Genentec, GlaxoSmithKline, Roche, TEVA, Takeda and Boehringer Ingelheim. KS has a patent pending for the Boston University: “Biomarkers of COPD disease activity”. IMB, AF, EvdW, EDT, SJMH, NHTtH, CAB, IHH, MRJ, HGdB, JSV, HRP, WB, FvdE, KM, AS and VG have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- World Health Organization. Global health observatory (GHO) data; top 10 causes of death. http://www.who.int/gho/mortality_burden_disease/causes_death/top_10/en/ [Accessed August 2017].
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

