Metagenomics Reveals the Impact of Wastewater Treatment Plants on the Dispersal of Microorganisms and Genes in Aquatic Sediments
- PMID: 29269503
- PMCID: PMC5812944
- DOI: 10.1128/AEM.02168-17
Metagenomics Reveals the Impact of Wastewater Treatment Plants on the Dispersal of Microorganisms and Genes in Aquatic Sediments
Abstract
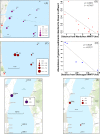
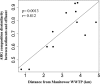
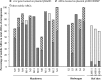
Wastewater treatment plants (WWTPs) release treated effluent containing mobile genetic elements (MGEs), antibiotic resistance genes (ARGs), and microorganisms into the environment, yet little is known about their influence on nearby microbial communities and the retention of these factors in receiving water bodies. Our research aimed to characterize the genes and organisms from two different WWTPs that discharge into Lake Michigan, as well as from surrounding lake sediments to determine the dispersal and fate of these factors with respect to distance from the effluent outfall. Shotgun metagenomics coupled to distance-decay analyses showed a higher abundance of genes identical to those in WWTP effluent genes in sediments closer to outfall sites than in sediments farther away, indicating their possible WWTP origin. We also found genes attributed to organisms, such as those belonging to Helicobacteraceae, Legionellaceae, Moraxellaceae, and Neisseriaceae, in effluent from both WWTPs and decreasing in abundance in lake sediments with increased distance from WWTPs. Moreover, our results showed that the WWTPs likely influence the ARG composition in lake sediments close to the effluent discharge. Many of these ARGs were located on MGEs in both the effluent and sediment samples, indicating a relatively broad propensity for horizontal gene transfer (HGT). Our approach allowed us to specifically link genes to organisms and their genetic context, providing insight into WWTP impacts on natural microbial communities. Overall, our results suggest a substantial influence of wastewater effluent on gene content and microbial community structure in the sediments of receiving water bodies.IMPORTANCE Wastewater treatment plants (WWTPs) release their effluent into aquatic environments. Although treated, effluent retains many genes and microorganisms that have the potential to influence the receiving water in ways that are poorly understood. Here, we tracked the genetic footprint, including genes specific to antibiotic resistance and mobile genetic elements and their associated organisms, from WWTPs to lake sediments. Our work is novel in that we used metagenomic data sets to comprehensively evaluate total gene content and the genetic and taxonomic context of specific genes in environmental samples putatively impacted by WWTP inputs. Based on two different WWTPs with different treatment processes, our findings point to an influence of WWTPs on the presence, abundance, and composition of these factors in the environment.
Keywords: antibiotic resistance; freshwater; lakes; metagenomics; wastewater treatment.
Copyright © 2018 Chu et al.
Figures

References
-
- Rizzo L, Mania C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. 2013. Urban wastewater treatment plant as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

