Cryo-EM structure of a licensed DNA replication origin
- PMID: 29269875
- PMCID: PMC5740162
- DOI: 10.1038/s41467-017-02389-0
Cryo-EM structure of a licensed DNA replication origin
Abstract
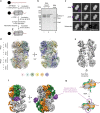
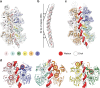
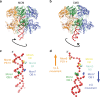
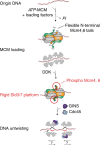
Eukaryotic origins of replication are licensed upon loading of the MCM helicase motor onto DNA. ATP hydrolysis by MCM is required for loading and the post-catalytic MCM is an inactive double hexamer that encircles duplex DNA. Origin firing depends on MCM engagement of Cdc45 and GINS to form the CMG holo-helicase. CMG assembly requires several steps including MCM phosphorylation by DDK. To understand origin activation, here we have determined the cryo-EM structures of DNA-bound MCM, either unmodified or phosphorylated, and visualize a phospho-dependent MCM element likely important for Cdc45 recruitment. MCM pore loops touch both the Watson and Crick strands, constraining duplex DNA in a bent configuration. By comparing our new MCM-DNA structure with the structure of CMG-DNA, we suggest how the conformational transition from the loaded, post-catalytic MCM to CMG might promote DNA untwisting and melting at the onset of replication.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

