Nanopore DNA Sequencing and Genome Assembly on the International Space Station
- PMID: 29269933
- PMCID: PMC5740133
- DOI: 10.1038/s41598-017-18364-0
Nanopore DNA Sequencing and Genome Assembly on the International Space Station
Abstract
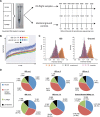
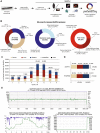
We evaluated the performance of the MinION DNA sequencer in-flight on the International Space Station (ISS), and benchmarked its performance off-Earth against the MinION, Illumina MiSeq, and PacBio RS II sequencing platforms in terrestrial laboratories. Samples contained equimolar mixtures of genomic DNA from lambda bacteriophage, Escherichia coli (strain K12, MG1655) and Mus musculus (female BALB/c mouse). Nine sequencing runs were performed aboard the ISS over a 6-month period, yielding a total of 276,882 reads with no apparent decrease in performance over time. From sequence data collected aboard the ISS, we constructed directed assemblies of the ~4.6 Mb E. coli genome, ~48.5 kb lambda genome, and a representative M. musculus sequence (the ~16.3 kb mitochondrial genome), at 100%, 100%, and 96.7% consensus pairwise identity, respectively; de novo assembly of the E. coli genome from raw reads yielded a single contig comprising 99.9% of the genome at 98.6% consensus pairwise identity. Simulated real-time analyses of in-flight sequence data using an automated bioinformatic pipeline and laptop-based genomic assembly demonstrated the feasibility of sequencing analysis and microbial identification aboard the ISS. These findings illustrate the potential for sequencing applications including disease diagnosis, environmental monitoring, and elucidating the molecular basis for how organisms respond to spaceflight.
Conflict of interest statement
Three authors (D.J.T., S.J. and F.I.) are employees of Oxford Nanopore Technologies, the company that produces the MinION sequencing technology. They assisted with experiment planning and instrument testing for flight. Analyses of nanopore data were performed independently by the bi-coastal team of the Chiu and Mason labs and the scientists at Oxford Nanopore Technologies. C.Y.C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center and receives research support from Abbott Laboratories, Inc. All other authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

