Variations of Histone Modification Patterns: Contributions of Inter-plant Variability and Technical Factors
- PMID: 29270186
- PMCID: PMC5725443
- DOI: 10.3389/fpls.2017.02084
Variations of Histone Modification Patterns: Contributions of Inter-plant Variability and Technical Factors
Abstract
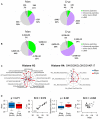
Inter-individual variability of conspecific plants is governed by differences in their genetically determined growth and development traits, environmental conditions, and adaptive responses under epigenetic control involving histone post-translational modifications. The apparent variability in histone modifications among plants might be increased by technical variation introduced in sample processing during epigenetic analyses. Thus, to detect true variations in epigenetic histone patterns associated with given factors, the basal variability among samples that is not associated with them must be estimated. To improve knowledge of relative contribution of biological and technical variation, mass spectrometry was used to examine histone modification patterns (acetylation and methylation) among Arabidopsis thaliana plants of ecotypes Columbia 0 (Col-0) and Wassilewskija (Ws) homogenized by two techniques (grinding in a cryomill or with a mortar and pestle). We found little difference in histone modification profiles between the ecotypes. However, in comparison of the biological and technical components of variability, we found consistently higher inter-individual variability in histone mark levels among Ws plants than among Col-0 plants (grown from seeds collected either from single plants or sets of plants). Thus, more replicates of Ws would be needed for rigorous analysis of epigenetic marks. Regarding technical variability, the cryomill introduced detectably more heterogeneity in the data than the mortar and pestle treatment, but mass spectrometric analyses had minor apparent effects. Our study shows that it is essential to consider inter-sample variance and estimate suitable numbers of biological replicates for statistical analysis for each studied organism when investigating changes in epigenetic histone profiles.
Keywords: Arabidopsis thaliana; ecotype; epigenetics; histone; mass spectrometry; post-translational modifications.
Figures



References
-
- Al Shweiki M. R., Mönchgesang S., Majovsky P., Thieme D., Trutschel D., Hoehenwarter W. (2017). Assessment of label-free quantification in discovery proteomics and impact of technological factors and natural variability of protein abundance. J. Proteome Res. 16 1410–1424. 10.1021/acs.jproteome.6b00645 - DOI - PubMed
-
- Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67 1–48. 10.18637/jss.v067.i01 - DOI
-
- Bennett K. L., Wang X., Bystrom C. E., Chambers M. C., Andacht T. M., Dangott L. J., et al. (2015). The 2012/2013 ABRF proteomic research group study: assessing longitudinal intralaboratory variability in routine peptide liquid chromatography tandem mass spectrometry analyses. Mol. Cell. Proteomics 14 3299–3309. 10.1074/mcp.O115.051888 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

