Cluster ensemble based on Random Forests for genetic data
- PMID: 29270227
- PMCID: PMC5732374
- DOI: 10.1186/s13040-017-0156-2
Cluster ensemble based on Random Forests for genetic data
Abstract
Background: Clustering plays a crucial role in several application domains, such as bioinformatics. In bioinformatics, clustering has been extensively used as an approach for detecting interesting patterns in genetic data. One application is population structure analysis, which aims to group individuals into subpopulations based on shared genetic variations, such as single nucleotide polymorphisms. Advances in DNA sequencing technology have facilitated the obtainment of genetic datasets with exceptional sizes. Genetic data usually contain hundreds of thousands of genetic markers genotyped for thousands of individuals, making an efficient means for handling such data desirable.
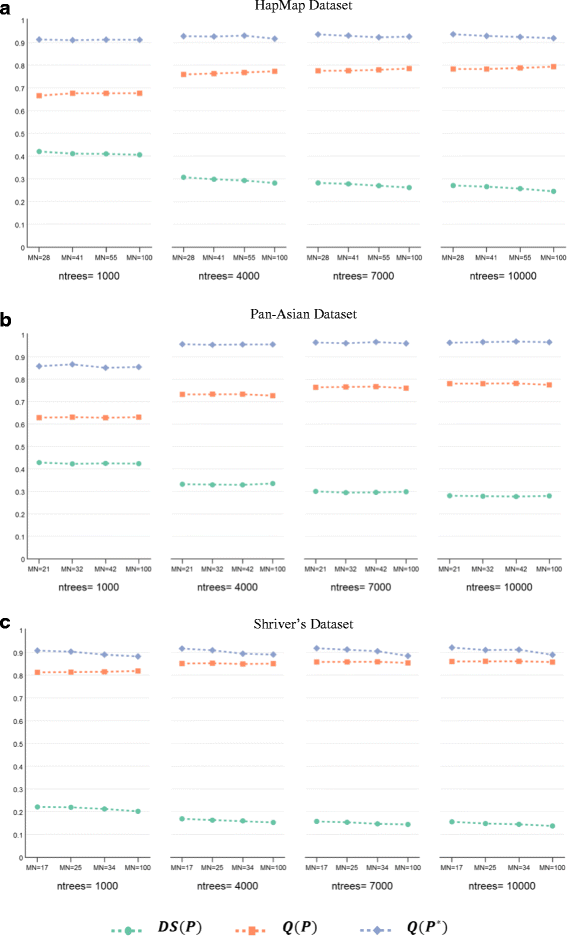
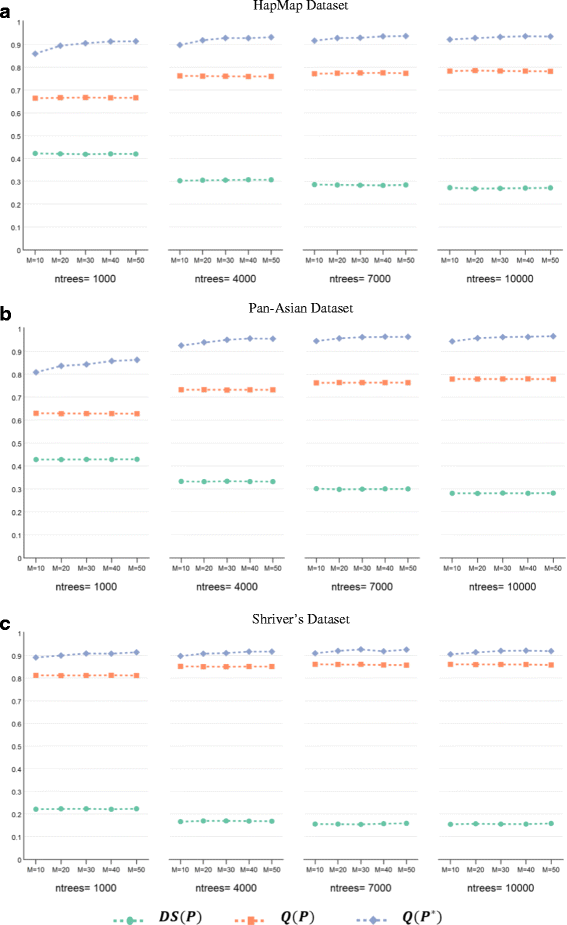
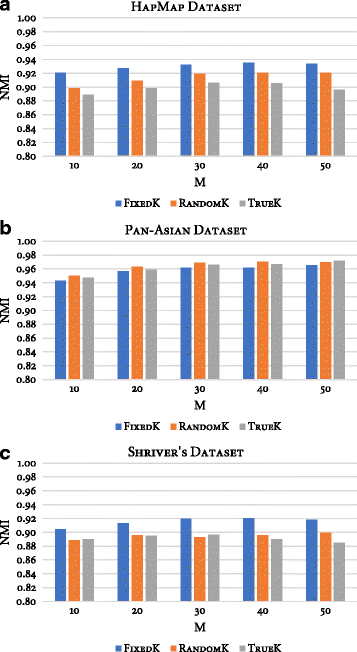
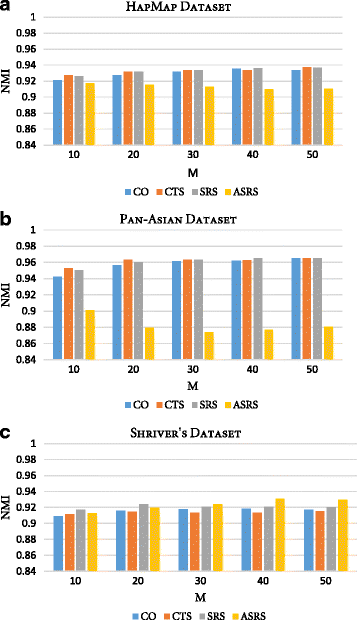
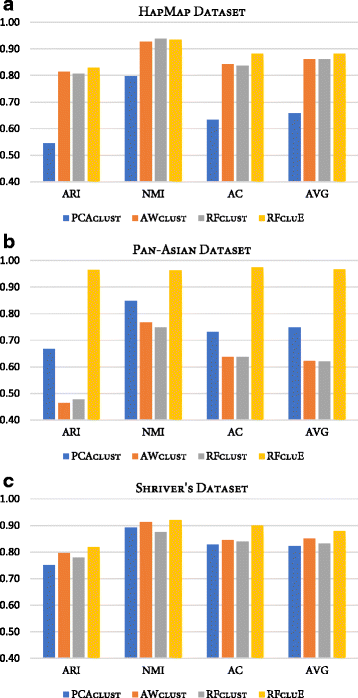
Results: Random Forests (RFs) has emerged as an efficient algorithm capable of handling high-dimensional data. RFs provides a proximity measure that can capture different levels of co-occurring relationships between variables. RFs has been widely considered a supervised learning method, although it can be converted into an unsupervised learning method. Therefore, RF-derived proximity measure combined with a clustering technique may be well suited for determining the underlying structure of unlabeled data. This paper proposes, RFcluE, a cluster ensemble approach for determining the underlying structure of genetic data based on RFs. The approach comprises a cluster ensemble framework to combine multiple runs of RF clustering. Experiments were conducted on high-dimensional, real genetic dataset to evaluate the proposed approach. The experiments included an examination of the impact of parameter changes, comparing RFcluE performance against other clustering methods, and an assessment of the relationship between the diversity and quality of the ensemble and its effect on RFcluE performance.
Conclusions: This paper proposes, RFcluE, a cluster ensemble approach based on RF clustering to address the problem of population structure analysis and demonstrate the effectiveness of the approach. The paper also illustrates that applying a cluster ensemble approach, combining multiple RF clusterings, produces more robust and higher-quality results as a consequence of feeding the ensemble with diverse views of high-dimensional genetic data obtained through bagging and random subspace, the two key features of the RF algorithm.
Keywords: Cluster ensemble; Ensemble diversity; Genetic population; High-dimensional data; Normalized mutual information; Population structure analysis; Random Forest proximity; Random Forests; Single nucleotide polymorphism.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

