Protein biomarkers for subtyping breast cancer and implications for future research
- PMID: 29271260
- PMCID: PMC6104835
- DOI: 10.1080/14789450.2018.1421071
Protein biomarkers for subtyping breast cancer and implications for future research
Abstract
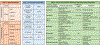
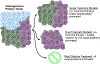
Breast cancer subtypes are currently defined by a combination of morphologic, genomic, and proteomic characteristics. These subtypes provide a molecular portrait of the tumor that aids diagnosis, prognosis, and treatment escalation/de-escalation options. Gene expression signatures describing intrinsic breast cancer subtypes for predicting risk of recurrence have been rapidly adopted in the clinic. Despite the use of subtype classifications, many patients develop drug resistance, breast cancer recurrence, or therapy failure. Areas covered: This review provides a summary of immunohistochemistry, reverse phase protein array, mass spectrometry, and integrative studies that are revealing differences in biological functions within and between breast cancer subtypes. We conclude with a discussion of rigor and reproducibility for proteomic-based biomarker discovery. Expert commentary: Innovations in proteomics, including implementation of assay guidelines and standards, are facilitating refinement of breast cancer subtypes. Proteomic and phosphoproteomic information distinguish biologically functional subtypes, are predictive of recurrence, and indicate likelihood of drug resistance. Actionable, activated signal transduction pathways can now be quantified and characterized. Proteomic biomarker validation in large, well-designed studies should become a public health priority to capitalize on the wealth of information gleaned from the proteome.
Keywords: Basal-like; HER2; biomarker; breast cancer; estrogen receptor; mass spectrometry; progesterone receptor; reverse phase protein array; signal transduction; triple negative breast cancer.
Figures


References
-
-
Malhotra GK, Zhao X, Band H, et al. Histological, molecular and functional subtypes of breast cancers. Cancer Biol Ther. 2010;10(10):955–60.
*Provides very good descriptions of breast histomorphology.
-
-
- Vasconcelos I, Hussainzada A, Berger S, et al. The St. Gallen surrogate classification for breast cancer subtypes successfully predicts tumor presenting features, nodal involvement, recurrence patterns and disease free survival. Breast. 2016;29:181–5. - PubMed
-
- Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction. The Journal of Pathology. 2010;220:263–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
