Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in Red and Green Walnut (Juglans regia L.)
- PMID: 29271948
- PMCID: PMC5943948
- DOI: 10.3390/molecules23010025
Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in Red and Green Walnut (Juglans regia L.)
Abstract
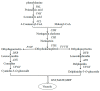
Fruit color is an important economic trait. The color of red walnut cultivars is mainly attributed to anthocyanins. The aim of this study was to explore the differences in the molecular mechanism of leaf and peel color change between red and green walnut. A reference transcriptome of walnut was sequenced and annotated to identify genes related to fruit color at the ripening stage. More than 290 million high-quality reads were assembled into 39,411 genes using a combined assembly strategy. Using Illumina digital gene expression profiling, we identified 4568 differentially expressed genes (DEGs) between red and green walnut leaf and 3038 DEGs between red and green walnut peel at the ripening stage. We also identified some transcription factor families (MYB, bHLH, and WD40) involved in the control of anthocyanin biosynthesis. The trends in the expression levels of several genes encoding anthocyanin biosynthetic enzymes and transcription factors in the leaf and peel of red and green walnut were verified by quantitative real-time PCR. Together, our results identified the genes involved in anthocyanin accumulation in red walnut. These data provide a valuable resource for understanding the coloration of red walnut.
Keywords: anthocyanins; differentially expressed genes (DEGs); leaf and peel; quantitative real-time PCR (qRT-PCR); red walnut; transcriptome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Aradhya M., Woeste K., Velasco D. Genetic diversity, structure and differentiation in cultivated walnut (Juglans regia L.) Acta Hortic. 2010;861:127–132. doi: 10.17660/ActaHortic.2010.861.16. - DOI
-
- Xi R.T., Zhang Y.P. Chinese Fruit Annals. Walnut. 1st ed. China Forestry Publishing House; Beijing, China: 1996. pp. 1–9.
-
- Mc Granahan G., Leslie C. ‘Robert Livermore’, a Persian walnut cultivar with a red seedcoat. Hortic. Sci. 2004;39:1772.
-
- Hanbali L.M.B., Ghadieh R.A., Hasan H.K., Nakhal Y.J., Haddad J. Measurement of antioxidant activity and antioxidant compounds under versatile extraction conditions: I. The immuno-biochemical antioxidant properties of sweet cherry (Prunus avium) extracts. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2013;12:173–187. doi: 10.2174/1871523011312020009. - DOI - PubMed
-
- Kelebek H., Selli S. Evaluation of chemical constituents and antioxidant activity of sweet cherry (Prunus avium L.) cultivars. Int. J. Food Sci. Technol. 2011;46:2530–2537. doi: 10.1111/j.1365-2621.2011.02777.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

