GIMDA: Graphlet interaction-based MiRNA-disease association prediction
- PMID: 29272076
- PMCID: PMC5824414
- DOI: 10.1111/jcmm.13429
GIMDA: Graphlet interaction-based MiRNA-disease association prediction
Abstract
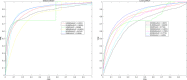
MicroRNAs (miRNAs) have been confirmed to be closely related to various human complex diseases by many experimental studies. It is necessary and valuable to develop powerful and effective computational models to predict potential associations between miRNAs and diseases. In this work, we presented a prediction model of Graphlet Interaction for MiRNA-Disease Association prediction (GIMDA) by integrating the disease semantic similarity, miRNA functional similarity, Gaussian interaction profile kernel similarity and the experimentally confirmed miRNA-disease associations. The related score of a miRNA to a disease was calculated by measuring the graphlet interactions between two miRNAs or two diseases. The novelty of GIMDA lies in that we used graphlet interaction to analyse the complex relationships between two nodes in a graph. The AUCs of GIMDA in global and local leave-one-out cross-validation (LOOCV) turned out to be 0.9006 and 0.8455, respectively. The average result of five-fold cross-validation reached to 0.8927 ± 0.0012. In case study for colon neoplasms, kidney neoplasms and prostate neoplasms based on the database of HMDD V2.0, 45, 45, 41 of the top 50 potential miRNAs predicted by GIMDA were validated by dbDEMC and miR2Disease. Additionally, in the case study of new diseases without any known associated miRNAs and the case study of predicting potential miRNA-disease associations using HMDD V1.0, there were also high percentages of top 50 miRNAs verified by the experimental literatures.
Keywords: disease; graphlet interaction; miRNA; miRNA-disease association.
© 2017 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures


Similar articles
-
An improved random forest-based computational model for predicting novel miRNA-disease associations.BMC Bioinformatics. 2019 Dec 3;20(1):624. doi: 10.1186/s12859-019-3290-7. BMC Bioinformatics. 2019. PMID: 31795954 Free PMC article.
-
EGBMMDA: Extreme Gradient Boosting Machine for MiRNA-Disease Association prediction.Cell Death Dis. 2018 Jan 5;9(1):3. doi: 10.1038/s41419-017-0003-x. Cell Death Dis. 2018. PMID: 29305594 Free PMC article.
-
MDHGI: Matrix Decomposition and Heterogeneous Graph Inference for miRNA-disease association prediction.PLoS Comput Biol. 2018 Aug 24;14(8):e1006418. doi: 10.1371/journal.pcbi.1006418. eCollection 2018 Aug. PLoS Comput Biol. 2018. PMID: 30142158 Free PMC article.
-
Review of MiRNA-Disease Association Prediction.Curr Protein Pept Sci. 2020;21(11):1044-1053. doi: 10.2174/1389203721666200210102751. Curr Protein Pept Sci. 2020. PMID: 32039677 Review.
-
MicroRNAs and complex diseases: from experimental results to computational models.Brief Bioinform. 2019 Mar 22;20(2):515-539. doi: 10.1093/bib/bbx130. Brief Bioinform. 2019. PMID: 29045685 Review.
Cited by
-
Self-Weighted Multi-Kernel Multi-Label Learning for Potential miRNA-Disease Association Prediction.Mol Ther Nucleic Acids. 2019 Sep 6;17:414-423. doi: 10.1016/j.omtn.2019.06.014. Epub 2019 Jun 28. Mol Ther Nucleic Acids. 2019. PMID: 31319245 Free PMC article.
-
MCLPMDA: A novel method for miRNA-disease association prediction based on matrix completion and label propagation.J Cell Mol Med. 2019 Feb;23(2):1427-1438. doi: 10.1111/jcmm.14048. Epub 2018 Nov 29. J Cell Mol Med. 2019. PMID: 30499204 Free PMC article.
-
MSFSP: A Novel miRNA-Disease Association Prediction Model by Federating Multiple-Similarities Fusion and Space Projection.Front Genet. 2020 Apr 30;11:389. doi: 10.3389/fgene.2020.00389. eCollection 2020. Front Genet. 2020. PMID: 32425980 Free PMC article.
-
FKL-Spa-LapRLS: an accurate method for identifying human microRNA-disease association.BMC Genomics. 2018 Dec 31;19(Suppl 10):911. doi: 10.1186/s12864-018-5273-x. BMC Genomics. 2018. PMID: 30598109 Free PMC article.
-
Identifying and Exploiting Potential miRNA-Disease Associations With Neighborhood Regularized Logistic Matrix Factorization.Front Genet. 2018 Aug 7;9:303. doi: 10.3389/fgene.2018.00303. eCollection 2018. Front Genet. 2018. PMID: 30131824 Free PMC article.
References
-
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001; 107: 823–6. - PubMed
-
- Meister G, Tuschl T. Mechanisms of gene silencing by double‐stranded RNA. Nature. 2004; 431: 343–9. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–97. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004; 431: 350–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

