Improved Precursor Characterization for Data-Dependent Mass Spectrometry
- PMID: 29272103
- PMCID: PMC5803309
- DOI: 10.1021/acs.analchem.7b04808
Improved Precursor Characterization for Data-Dependent Mass Spectrometry
Abstract
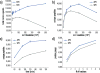
Modern ion trap mass spectrometers are capable of collecting up to 60 tandem MS (MS/MS) scans per second, in theory providing acquisition speeds that can sample every eluting peptide precursor presented to the MS system. In practice, however, the precursor sampling capacity enabled by these ultrafast acquisition rates is often underutilized due to a host of reasons (e.g., long injection times and wide analyzer mass ranges). One often overlooked reason for this underutilization is that the instrument exhausts all the peptide features it identifies as suitable for MS/MS fragmentation. Highly abundant features can prevent annotation of lower abundance precursor ions that occupy similar mass-to-charge (m/z) space, which ultimately inhibits the acquisition of an MS/MS event. Here, we present an advanced peak determination (APD) algorithm that uses an iterative approach to annotate densely populated m/z regions to increase the number of peptides sampled during data-dependent LC-MS/MS analyses. The APD algorithm enables nearly full utilization of the sampling capacity of a quadrupole-Orbitrap-linear ion trap MS system, which yields up to a 40% increase in unique peptide identifications from whole cell HeLa lysates (approximately 53 000 in a 90 min LC-MS/MS analysis). The APD algorithm maintains improved peptide and protein identifications across several modes of proteomic data acquisition, including varying gradient lengths, different degrees of prefractionation, peptides derived from multiple proteases, and phosphoproteomic analyses. Additionally, the use of APD increases the number of peptides characterized per protein, providing improved protein quantification. In all, the APD algorithm increases the number of detectable peptide features, which maximizes utilization of the high MS/MS capacities and significantly improves sampling depth and identifications in proteomic experiments.
Figures

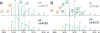




Similar articles
-
Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes.Mol Cell Proteomics. 2012 Mar;11(3):O111.013698. doi: 10.1074/mcp.O111.013698. Epub 2011 Dec 9. Mol Cell Proteomics. 2012. PMID: 22159718 Free PMC article.
-
Advanced Precursor Ion Selection Algorithms for Increased Depth of Bottom-Up Proteomic Profiling.J Proteome Res. 2016 Oct 7;15(10):3563-3573. doi: 10.1021/acs.jproteome.6b00312. Epub 2016 Sep 7. J Proteome Res. 2016. PMID: 27569903 Free PMC article.
-
Variable-Velocity Traveling-Wave Ion Mobility Separation Enhancing Peak Capacity for Data-Independent Acquisition Proteomics.Anal Chem. 2017 Jun 6;89(11):5669-5672. doi: 10.1021/acs.analchem.7b00112. Epub 2017 May 9. Anal Chem. 2017. PMID: 28471653 Free PMC article.
-
High-throughput database search and large-scale negative polarity liquid chromatography-tandem mass spectrometry with ultraviolet photodissociation for complex proteomic samples.Mol Cell Proteomics. 2013 Sep;12(9):2604-14. doi: 10.1074/mcp.O113.028258. Epub 2013 May 21. Mol Cell Proteomics. 2013. PMID: 23695934 Free PMC article.
-
Multiplexed and data-independent tandem mass spectrometry for global proteome profiling.Mass Spectrom Rev. 2014 Nov-Dec;33(6):452-70. doi: 10.1002/mas.21400. Epub 2013 Nov 26. Mass Spectrom Rev. 2014. PMID: 24281846 Review.
Cited by
-
High-Throughput, Comprehensive Single-Cell Proteomic Analysis of Xenopus laevis Embryos at the 50-Cell Stage Using a Microplate-Based MICROFASP System.Anal Chem. 2022 Feb 22;94(7):3254-3259. doi: 10.1021/acs.analchem.1c04987. Epub 2022 Feb 10. Anal Chem. 2022. PMID: 35143156 Free PMC article.
-
Protein Contaminants Matter: Building Universal Protein Contaminant Libraries for DDA and DIA Proteomics.J Proteome Res. 2022 Sep 2;21(9):2104-2113. doi: 10.1021/acs.jproteome.2c00145. Epub 2022 Jul 6. J Proteome Res. 2022. PMID: 35793413 Free PMC article.
-
Large-scale Multi-omic Analysis of COVID-19 Severity.medRxiv [Preprint]. 2020 Jul 19:2020.07.17.20156513. doi: 10.1101/2020.07.17.20156513. medRxiv. 2020. Update in: Cell Syst. 2021 Jan 20;12(1):23-40.e7. doi: 10.1016/j.cels.2020.10.003. PMID: 32743614 Free PMC article. Updated. Preprint.
-
Metabolic Remodeling during Nitrogen Fixation in Zymomonas mobilis.mSystems. 2021 Dec 21;6(6):e0098721. doi: 10.1128/mSystems.00987-21. Epub 2021 Nov 16. mSystems. 2021. PMID: 34783580 Free PMC article.
-
Constructing and deconstructing GATA2-regulated cell fate programs to establish developmental trajectories.J Exp Med. 2020 Nov 2;217(11):e20191526. doi: 10.1084/jem.20191526. J Exp Med. 2020. PMID: 32736380 Free PMC article.
References
-
- Washburn MP, Wolters D, Yates JR., III Nature biotechnology. 2001;19:242. - PubMed
-
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Journal of Proteome Research. 2003;2:43–50. - PubMed
-
- Caprioli RM, Farmer TB, Gile J. Analytical Chemistry. 1997;69:4751–4760. - PubMed
-
- Wollnik H, Przewloka M. Int. J. Mass Spectrom. Ion Process. 1990;96:267–274.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

