A survey of localized sequence rearrangements in human DNA
- PMID: 29272440
- PMCID: PMC5829575
- DOI: 10.1093/nar/gkx1266
A survey of localized sequence rearrangements in human DNA
Abstract
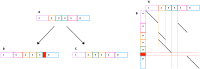
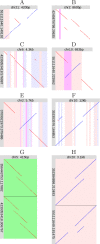
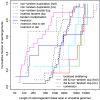
Genomes mutate and evolve in ways simple (substitution or deletion of bases) and complex (e.g. chromosome shattering). We do not fully understand what types of complex mutation occur, and we cannot routinely characterize arbitrarily-complex mutations in a high-throughput, genome-wide manner. Long-read DNA sequencing methods (e.g. PacBio, nanopore) are promising for this task, because one read may encompass a whole complex mutation. We describe an analysis pipeline to characterize arbitrarily-complex 'local' mutations, i.e. intrachromosomal mutations encompassed by one DNA read. We apply it to nanopore and PacBio reads from one human cell line (NA12878), and survey sequence rearrangements, both real and artifactual. Almost all the real rearrangements belong to recurring patterns or motifs: the most common is tandem multiplication (e.g. heptuplication), but there are also complex patterns such as localized shattering, which resembles DNA damage by radiation. Gene conversions are identified, including one between hemoglobin gamma genes. This study demonstrates a way to find intricate rearrangements with any number of duplications, deletions, and repositionings. It demonstrates a probability-based method to resolve ambiguous rearrangements involving highly similar sequences, as occurs in gene conversion. We present a catalog of local rearrangements in one human cell line, and show which rearrangement patterns occur.
Figures



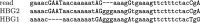
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

