Triggering nucleic acid nanostructure assembly by conditional kissing interactions
- PMID: 29272518
- PMCID: PMC5814900
- DOI: 10.1093/nar/gkx1267
Triggering nucleic acid nanostructure assembly by conditional kissing interactions
Abstract
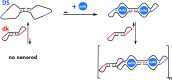
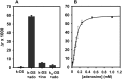
Nucleic acids are biomolecules of amazing versatility. Beyond their function for information storage they can be used for building nano-objects. We took advantage of loop-loop or kissing interactions between hairpin building blocks displaying complementary loops for driving the assembly of nucleic acid nano-architectures. It is of interest to make the interaction between elementary units dependent on an external trigger, thus allowing the control of the scaffold formation. To this end we exploited the binding properties of structure-switching aptamers (aptaswitch). Aptaswitches are stem-loop structured oligonucleotides that engage a kissing complex with an RNA hairpin in response to ligand-induced aptaswitch folding. We demonstrated the potential of this approach by conditionally assembling oligonucleotide nanorods in response to the addition of adenosine.
Figures



References
-
- Chandrasekaran A.R. Programmable DNA scaffolds for spatially-ordered protein assembly. Nanoscale. 2016; 8:4436–4446. - PubMed
-
- Seeman N.C. DNA in a material world. Nature. 2003; 421:427–431. - PubMed
-
- Seeman N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982; 99:237–247. - PubMed
-
- Chen J., Seeman N.C.. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991; 350:631–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

