Genomic and Proteomic Resolution of Heterochromatin and Its Restriction of Alternate Fate Genes
- PMID: 29272703
- PMCID: PMC5858919
- DOI: 10.1016/j.molcel.2017.11.030
Genomic and Proteomic Resolution of Heterochromatin and Its Restriction of Alternate Fate Genes
Abstract
Heterochromatin is integral to cell identity maintenance by impeding the activation of genes for alternate cell fates. Heterochromatic regions are associated with histone 3 lysine 9 trimethylation (H3K9me3) or H3K27me3, but these modifications are also found in euchromatic regions that permit transcription. We discovered that resistance to sonication is a reliable indicator of the heterochromatin state, and we developed a biophysical method (gradient-seq) to discriminate subtypes of H3K9me3 and H3K27me3 domains in sonication-resistant heterochromatin (srHC) versus euchromatin. These classifications are more accurate than the histone marks alone in predicting transcriptional silence and resistance of alternate fate genes to activation during direct cell conversion. Our proteomics of H3K9me3-marked srHC and functional screens revealed diverse proteins, including RBMX and RBMXL1, that impede gene induction during cellular reprogramming. Isolation of srHC with gradient-seq provides a genome-wide map of chromatin structure, elucidating subtypes of repressed domains that are uniquely predictive of diverse other chromatin properties.
Keywords: DBRs; H3K27me3; H3K9me3; chromatin organization; gene repression; gradient-seq; heterochromatin; hiHep; proteomics; reprogramming.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
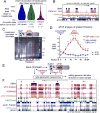


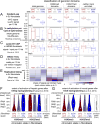
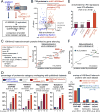

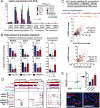
Comment in
-
Beyond the Histone Code: A Physical Map of Chromatin States.Mol Cell. 2018 Jan 4;69(1):5-7. doi: 10.1016/j.molcel.2017.12.018. Mol Cell. 2018. PMID: 29304334
References
-
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

