AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection
- PMID: 29272704
- PMCID: PMC5744881
- DOI: 10.1016/j.molcel.2017.11.026
AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection
Abstract
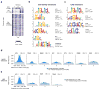
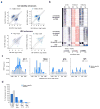
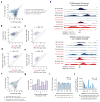
Enhancer elements are genomic regulatory sequences that direct the selective expression of genes so that genetically identical cells can differentiate and acquire the highly specialized forms and functions required to build a functioning animal. To differentiate, cells must select from among the ∼106 enhancers encoded in the genome the thousands of enhancers that drive the gene programs that impart their distinct features. We used a genetic approach to identify transcription factors (TFs) required for enhancer selection in fibroblasts. This revealed that the broadly expressed, growth-factor-inducible TFs FOS/JUN (AP-1) play a central role in enhancer selection. FOS/JUN selects enhancers together with cell-type-specific TFs by collaboratively binding to nucleosomal enhancers and recruiting the SWI/SNF (BAF) chromatin remodeling complex to establish accessible chromatin. These experiments demonstrate how environmental signals acting via FOS/JUN and BAF coordinate with cell-type-specific TFs to select enhancer repertoires that enable differentiation during development.
Keywords: Ras/MAPK signaling; chromatin remodeling complexes; enhancers; genetics; genomics; growth factor signaling; lineage specification; mSWI/SNF (BAF) complexes; transcription factors; transcriptional regulation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

