The two TRAPP complexes of metazoans have distinct roles and act on different Rab GTPases
- PMID: 29273580
- PMCID: PMC5800803
- DOI: 10.1083/jcb.201705068
The two TRAPP complexes of metazoans have distinct roles and act on different Rab GTPases
Abstract
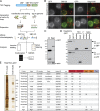
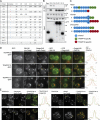
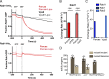
Originally identified in yeast, transport protein particle (TRAPP) complexes are Rab GTPase exchange factors that share a core set of subunits. TRAPPs were initially found to act on Ypt1, the yeast orthologue of Rab1, but recent studies have found that yeast TRAPPII can also activate the Rab11 orthologues Ypt31/32. Mammals have two TRAPP complexes, but their role is less clear, and they contain subunits that are not found in the yeast complexes but are essential for cell growth. To investigate TRAPP function in metazoans, we show that Drosophila melanogaster have two TRAPP complexes similar to those in mammals and that both activate Rab1, whereas one, TRAPPII, also activates Rab11. TRAPPII is not essential but becomes so in the absence of the gene parcas that encodes the Drosophila orthologue of the SH3BP5 family of Rab11 guanine nucleotide exchange factors (GEFs). Thus, in metazoans, Rab1 activation requires TRAPP subunits not found in yeast, and Rab11 activation is shared by TRAPPII and an unrelated GEF that is metazoan specific.
© 2018 MRC Laboratory of Molecular Biology.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

