RecQ and Fe-S helicases have unique roles in DNA metabolism dictated by their unwinding directionality, substrate specificity, and protein interactions
- PMID: 29273621
- PMCID: PMC5863537
- DOI: 10.1042/BST20170044
RecQ and Fe-S helicases have unique roles in DNA metabolism dictated by their unwinding directionality, substrate specificity, and protein interactions
Abstract
Helicases are molecular motors that play central roles in nucleic acid metabolism. Mutations in genes encoding DNA helicases of the RecQ and iron-sulfur (Fe-S) helicase families are linked to hereditary disorders characterized by chromosomal instabilities, highlighting the importance of these enzymes. Moreover, mono-allelic RecQ and Fe-S helicase mutations are associated with a broad spectrum of cancers. This review will discuss and contrast the specialized molecular functions and biological roles of RecQ and Fe-S helicases in DNA repair, the replication stress response, and the regulation of gene expression, laying a foundation for continued research in these important areas of study.
Keywords: DNA replication and recombination; DNA synthesis and repair; cancer; genetic disease; genome integrity; helicase.
© 2018 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures


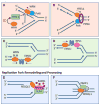
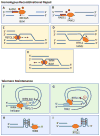
References
-
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. https://doi.org/10.1146/annurev.biochem.76.052305.115300. - DOI - PubMed
-
- Kaiser S, Sauer F, Kisker C. The structural and functional characterization of human RecQ4 reveals insights into its helicase mechanism. Nat Commun. 2017;8:15907. https://doi.org/10.1038/ncomms15907. - DOI - PMC - PubMed
-
- Newman JA, Aitkenhead H, Savitsky P, Gileadi O. Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5. Nucleic Acids Res. 2017;45:4231–4243. https://doi.org/10.1093/nar/gkw1362. - DOI - PMC - PubMed
-
- Manthei KA, Hill MC, Burke JE, Butcher SE, Keck JL. Structural mechanisms of DNA binding and unwinding in bacterial RecQ helicases. Proc Natl Acad Sci USA. 2015;112:4292–4297. https://doi.org/10.1073/pnas.1416746112. - DOI - PMC - PubMed
-
- Newman JA, Savitsky P, Allerston CK, Bizard AH, Özer Ö, Sarlós K, et al. Crystal structure of the Bloom’s syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids Res. 2015;43:5221–5235. https://doi.org/10.1093/nar/gkv373. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

