Topographical diversity of common skin microflora and its association with skin environment type: An observational study in Chinese women
- PMID: 29273721
- PMCID: PMC5741767
- DOI: 10.1038/s41598-017-18181-5
Topographical diversity of common skin microflora and its association with skin environment type: An observational study in Chinese women
Abstract
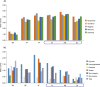
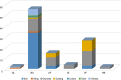
This study evaluated cutaneous microbial distribution, and microbial co-occurrence at different body sites and skin environments in Chinese women (39.6 ± 11.9 years, N = 100) during the winter season. Microbial distribution (Propionibacterium acnes, Staphylococcus aureus, Staphylococcus epidermidis, Lactobacillus, Pseudomonadaceae, and Malassezia furfur), association with biomarkers (antimicrobial peptides: LL-37, β-defensins [HBD-2, HBD-3]), and claudin-1) and skin biophysical parameters (transepidermal water loss, pH, skin scaliness and roughness, sebum and hydration levels) were also determined. Skin sites (glabella [GL], hand-back [HB], interdigital web-space [IS], antecubital fossa [AF], volar forearm [VF], back [BA]) were classified as normal, oily or dry based on two-step cluster analysis and exposed or unexposed (uncovered or covered by clothes, respectively) based on seasonal apparel. Pseudomonadaceae and Staphylococcus aureus had the highest and lowest detection rate respectively at all sites. Cluster analysis identified skin sites as 'normal' (HB, BA, AF, VF), 'dry' (IS) and 'oily' (GL). Bacterial alpha diversity was higher in exposed (HB, IS, and GL) compared with unexposed sites (BA, AF and VF). Co-occurrence of Staphylococcus aureus with any of the other five microorganisms was lower in dry and oily skin versus normal skin. Skin exposure, biophysical/barrier profile and biomarkers were found to be associated with bacterial distribution and co-occurrence.
Conflict of interest statement
X.L. and L.X. are employees of Johnson & Johnson (China) Ltd. C.Y. and P.H. have received grant from Johnson & Johnson Ltd. This study was funded by Johnson & Johnson (Asia Pacific).
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

