Alternative dominance of the parental genomes in hybrid cells generated through the fusion of mouse embryonic stem cells with fibroblasts
- PMID: 29273752
- PMCID: PMC5741742
- DOI: 10.1038/s41598-017-18352-4
Alternative dominance of the parental genomes in hybrid cells generated through the fusion of mouse embryonic stem cells with fibroblasts
Abstract
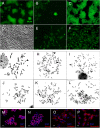
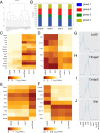
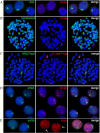
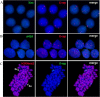
For the first time, two types of hybrid cells with embryonic stem (ES) cell-like and fibroblast-like phenotypes were produced through the fusion of mouse ES cells with fibroblasts. Transcriptome analysis of 2,848 genes differentially expressed in the parental cells demonstrated that 34-43% of these genes are expressed in hybrid cells, consistent with their phenotypes; 25-29% of these genes display intermediate levels of expression, and 12-16% of these genes maintained expression at the parental cell level, inconsistent with the phenotype of the hybrid cell. Approximately 20% of the analyzed genes displayed unexpected expression patterns that differ from both parents. An unusual phenomenon was observed, namely, the illegitimate activation of Xist expression and the inactivation of one of two X-chromosomes in the near-tetraploid fibroblast-like hybrid cells, whereas both Xs were active before and after in vitro differentiation of the ES cell-like hybrid cells. These results and previous data obtained on heterokaryons suggest that the appearance of hybrid cells with a fibroblast-like phenotype reflects the reprogramming, rather than the induced differentiation, of the ES cell genome under the influence of a somatic partner.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

