Amino acid residues in HIV-2 reverse transcriptase that restrict the development of nucleoside analogue resistance through the excision pathway
- PMID: 29275329
- PMCID: PMC5818179
- DOI: 10.1074/jbc.RA117.000177
Amino acid residues in HIV-2 reverse transcriptase that restrict the development of nucleoside analogue resistance through the excision pathway
Abstract
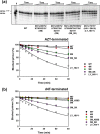
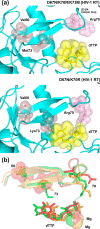
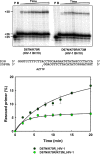
Nucleoside reverse transcriptase (RT) inhibitors (NRTIs) are the backbone of current antiretroviral treatments. However, the emergence of viral resistance against NRTIs is a major threat to their therapeutic effectiveness. In HIV-1, NRTI resistance-associated mutations either reduce RT-mediated incorporation of NRTI triphosphates (discrimination mechanism) or confer an ATP-mediated nucleotide excision activity that removes the inhibitor from the 3' terminus of DNA primers, enabling further primer elongation (excision mechanism). In HIV-2, resistance to zidovudine (3'-azido-3'-deoxythymidine (AZT)) and other NRTIs is conferred by mutations affecting nucleotide discrimination. Mutations of the excision pathway such as M41L, D67N, K70R, or S215Y (known as thymidine-analogue resistance mutations (TAMs)) are rare in the virus from HIV-2-infected individuals. Here, we demonstrate that mutant M41L/D67N/K70R/S215Y HIV-2 RT lacks ATP-dependent excision activity, and recombinant virus containing this RT remains susceptible to AZT inhibition. Mutant HIV-2 RTs were tested for their ability to unblock and extend DNA primers terminated with AZT and other NRTIs, when complexed with RNA or DNA templates. Our results show that Met73 and, to a lesser extent, Ile75 suppress excision activity when TAMs are present in the HIV-2 RT. Interestingly, recombinant HIV-2 carrying a mutant D67N/K70R/M73K RT showed 10-fold decreased AZT susceptibility and increased rescue efficiency on AZT- or tenofovir-terminated primers, as compared with the double-mutant D67N/K70R. Molecular dynamics simulations reveal that Met73influences β3-β4 hairpin loop conformation, whereas its substitution affects hydrogen bond interactions at position 70, required for NRTI excision. Our work highlights critical HIV-2 RT residues impeding the development of excision-mediated NRTI resistance.
Keywords: ATP-dependent excision; DNA replication; antiretroviral drugs; drug resistance; human immunodeficiency virus (HIV); nucleoside/nucleotide analogue; reverse transcription.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision.J Virol. 2002 Sep;76(18):9143-51. doi: 10.1128/jvi.76.18.9143-9151.2002. J Virol. 2002. PMID: 12186898 Free PMC article.
-
Effects of the Delta67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision.J Virol. 2004 Sep;78(18):9987-97. doi: 10.1128/JVI.78.18.9987-9997.2004. J Virol. 2004. PMID: 15331732 Free PMC article.
-
Molecular basis of the association of H208Y and thymidine analogue resistance mutations M41L, L210W and T215Y in the HIV-1 reverse transcriptase of treated patients.Antiviral Res. 2014 Jun;106:42-52. doi: 10.1016/j.antiviral.2014.03.004. Epub 2014 Mar 22. Antiviral Res. 2014. PMID: 24667336
-
Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure.Antivir Ther. 2001;6 Suppl 3:25-44. Antivir Ther. 2001. PMID: 11678471 Review.
-
Primer unblocking by HIV-1 reverse transcriptase and resistance to nucleoside RT inhibitors (NRTIs).Int J Biochem Cell Biol. 2004 Sep;36(9):1687-705. doi: 10.1016/j.biocel.2004.02.028. Int J Biochem Cell Biol. 2004. PMID: 15183338 Review.
Cited by
-
Performance evaluation of a laboratory developed PCR test for quantitation of HIV-2 viral RNA.PLoS One. 2020 Feb 28;15(2):e0229424. doi: 10.1371/journal.pone.0229424. eCollection 2020. PLoS One. 2020. PMID: 32109949 Free PMC article.
-
Antiretroviral Treatment of HIV-2 Infection: Available Drugs, Resistance Pathways, and Promising New Compounds.Int J Mol Sci. 2023 Mar 21;24(6):5905. doi: 10.3390/ijms24065905. Int J Mol Sci. 2023. PMID: 36982978 Free PMC article. Review.
-
Expanded Spectrum of Antiretroviral-Selected Mutations in Human Immunodeficiency Virus Type 2.J Infect Dis. 2020 Jun 11;221(12):1962-1972. doi: 10.1093/infdis/jiaa026. J Infect Dis. 2020. PMID: 31965175 Free PMC article.
-
Analysis and Molecular Determinants of HIV RNase H Cleavage Specificity at the PPT/U3 Junction.Viruses. 2021 Jan 18;13(1):131. doi: 10.3390/v13010131. Viruses. 2021. PMID: 33477685 Free PMC article.
-
Distinct Antiretroviral Mechanisms Elicited by a Viral Mutagen.J Mol Biol. 2021 Sep 3;433(18):167111. doi: 10.1016/j.jmb.2021.167111. Epub 2021 Jun 18. J Mol Biol. 2021. PMID: 34153286 Free PMC article.
References
-
- Smith R. A., Gottlieb G. S., Anderson D. J., Pyrak C. L., and Preston B. D. (2008) Human immunodeficiency virus types 1 and 2 exhibit comparable sensitivities to zidovudine and other nucleoside analog inhibitors in vitro. Antimicrob. Agents Chemother. 52, 329–332 10.1128/AAC.01004-07 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

