Cancer Immunotherapy Targets Based on Understanding the T Cell-Inflamed Versus Non-T Cell-Inflamed Tumor Microenvironment
- PMID: 29275462
- PMCID: PMC6693322
- DOI: 10.1007/978-3-319-67577-0_2
Cancer Immunotherapy Targets Based on Understanding the T Cell-Inflamed Versus Non-T Cell-Inflamed Tumor Microenvironment
Abstract
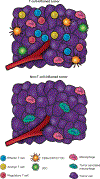
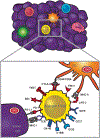
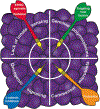
Most cancers express tumor antigens that can be recognized by T cells of the host. The fact that cancers become clinically evident nonetheless implies that immune escape must occur. Two major subsets of human melanoma metastases have been identified based on gene expression profiling. One subgroup has a T cell-inflamed phenotype that includes expression of chemokines, T cell markers, and a type I IFN signature. In contrast, the other major subset lacks this phenotype and has been designated as non-T cell-inflamed. The mechanisms of immune escape are likely distinct in these two phenotypes, and therefore the optimal immunotherapeutic interventions necessary to promote clinical responses may be different. The T cell-inflamed tumor microenvironment subset shows the highest expression of negative regulatory factors, including PD-L1, IDO, FoxP3+ Tregs, and evidence for T cell-intrinsic anergy. Therapeutic strategies to overcome these inhibitory mechanisms are being pursued, and anti-PD-1 mAbs have been FDA approved. The presence of multiple inhibitory mechanisms in the same tumor microenvironment argues that combination therapies may be advantageous, several of which are in clinical testing. A new paradigm may be needed to promote de novo inflammation in cases of the non-T cell-infiltrated tumor microenvironment. Natural innate immune sensing of tumors appears to occur via the host STING pathway, type I IFN production, and cross-priming of T cells via CD8α+ DCs. New strategies are being developed to engage this pathway therapeutically, such as through STING agonists. The molecular mechanisms that mediate the presence or absence of the T cell-inflamed tumor microenvironment are being elucidated using parallel genomics platforms. The first oncogene pathway identified that mediates immune exclusion is the Wnt/β-catenin pathway, suggesting that new pharmacologic strategies to target this pathway should be developed to restore immune access to the tumor microenvironment.
Keywords: Cancer immunotherapy; Checkpoint blockade; Dendritic cells; Immune evasion; Innate immune sensing; T cell dysfunction; T cell inflammation; Tumor microenvironment.
Figures
References
-
- Peterson AC, Harlin H, Gajewski TF. Immunization with melan-A peptide-pulsed peripheral blood mono-nuclear cells plus recombinant human interleukin-12 induces clinical activity and T-cell responses in advanced melanoma. J Clin Oncol. 2003;21:2342–8. - PubMed
-
- Gajewski TF, Meng Y, Blank C, Brown I, Kacha A, Kline J, Harlin H. Immune resistance orchestrated by the tumor microenvironment. Immunol Rev. 2006;213:131–45. - PubMed
-
- Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J. 2010;16:399–403. - PubMed
-
- Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF, Suciu S, Kruit WH, Eggermont AM, Vansteenkiste J, Brichard VG. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol. 2013;31:2388–95. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

