Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner
- PMID: 29275859
- PMCID: PMC5786503
- DOI: 10.1016/j.cell.2017.11.042
Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner
Abstract
Microglia are embryonically seeded macrophages that contribute to brain development, homeostasis, and pathologies. It is thus essential to decipher how microglial properties are temporally regulated by intrinsic and extrinsic factors, such as sexual identity and the microbiome. Here, we found that microglia undergo differentiation phases, discernable by transcriptomic signatures and chromatin accessibility landscapes, which can diverge in adult males and females. Remarkably, the absence of microbiome in germ-free mice had a time and sexually dimorphic impact both prenatally and postnatally: microglia were more profoundly perturbed in male embryos and female adults. Antibiotic treatment of adult mice triggered sexually biased microglial responses revealing both acute and long-term effects of microbiota depletion. Finally, human fetal microglia exhibited significant overlap with the murine transcriptomic signature. Our study shows that microglia respond to environmental challenges in a sex- and time-dependent manner from prenatal stages, with major implications for our understanding of microglial contributions to health and disease.
Keywords: CXCR4; antibiotics; embryogenesis; germ-free; microbiome; microglia; neurodevelopmental disorders; neuroinflammation; prenatal; sex.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
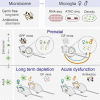
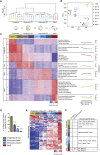

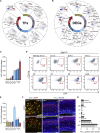


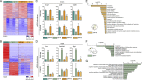




Comment in
-
Sex, Gut, and Microglia.Dev Cell. 2018 Jan 22;44(2):137-138. doi: 10.1016/j.devcel.2018.01.003. Dev Cell. 2018. PMID: 29401417
References
-
- Arnò B., Grassivaro F., Rossi C., Bergamaschi A., Castiglioni V., Furlan R., Greter M., Favaro R., Comi G., Becher B. Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat. Commun. 2014;5:5611. - PubMed
-
- Beaume M., Hernandez D., Docquier M., Delucinge-Vivier C., Descombes P., François P. Orientation and expression of methicillin-resistant Staphylococcus aureus small RNAs by direct multiplexed measurements using the nCounter of NanoString technology. J. Microbiol. Methods. 2011;84:327–334. - PubMed
-
- Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

