Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes
- PMID: 29275860
- PMCID: PMC5786495
- DOI: 10.1016/j.cell.2017.11.043
Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes
Abstract
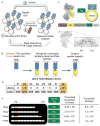
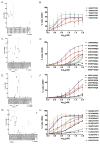
The immune system can mount T cell responses against tumors; however, the antigen specificities of tumor-infiltrating lymphocytes (TILs) are not well understood. We used yeast-display libraries of peptide-human leukocyte antigen (pHLA) to screen for antigens of "orphan" T cell receptors (TCRs) expressed on TILs from human colorectal adenocarcinoma. Four TIL-derived TCRs exhibited strong selection for peptides presented in a highly diverse pHLA-A∗02:01 library. Three of the TIL TCRs were specific for non-mutated self-antigens, two of which were present in separate patient tumors, and shared specificity for a non-mutated self-antigen derived from U2AF2. These results show that the exposed recognition surface of MHC-bound peptides accessible to the TCR contains sufficient structural information to enable the reconstruction of sequences of peptide targets for pathogenic TCRs of unknown specificity. This finding underscores the surprising specificity of TCRs for their cognate antigens and enables the facile indentification of tumor antigens through unbiased screening.
Keywords: T cell; T cell receptor; antigens; cancer; combinatorial biology; human leukocyte antigen; ligand identification; peptide library; peptides; single-cell sequencing.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





Comment in
-
What can I TIL you? Decoding TCR antigens.Sci Immunol. 2018 Feb 2;3(20):eaat0810. doi: 10.1126/sciimmunol.aat0810. Sci Immunol. 2018. PMID: 29429980
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI125197/AI/NIAID NIH HHS/United States
- K08 DK100739/DK/NIDDK NIH HHS/United States
- R01 AI103867/AI/NIAID NIH HHS/United States
- L30 AI113847/AI/NIAID NIH HHS/United States
- U19 AI109662/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- L30 DK089634/DK/NIDDK NIH HHS/United States
- R01 AI048540/AI/NIAID NIH HHS/United States
- K01 CA175127/CA/NCI NIH HHS/United States
- P01 CA132681/CA/NCI NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- F31 CA216926/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

