Missense Variants in RHOBTB2 Cause a Developmental and Epileptic Encephalopathy in Humans, and Altered Levels Cause Neurological Defects in Drosophila
- PMID: 29276004
- PMCID: PMC5777381
- DOI: 10.1016/j.ajhg.2017.11.008
Missense Variants in RHOBTB2 Cause a Developmental and Epileptic Encephalopathy in Humans, and Altered Levels Cause Neurological Defects in Drosophila
Abstract
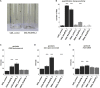
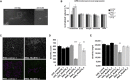
Although the role of typical Rho GTPases and other Rho-linked proteins in synaptic plasticity and cognitive function and dysfunction is widely acknowledged, the role of atypical Rho GTPases (such as RHOBTB2) in neurodevelopment has barely been characterized. We have now identified de novo missense variants clustering in the BTB-domain-encoding region of RHOBTB2 in ten individuals with a similar phenotype, including early-onset epilepsy, severe intellectual disability, postnatal microcephaly, and movement disorders. Three of the variants were recurrent. Upon transfection of HEK293 cells, we found that mutant RHOBTB2 was more abundant than the wild-type, most likely because of impaired degradation in the proteasome. Similarly, elevated amounts of the Drosophila ortholog RhoBTB in vivo were associated with seizure susceptibility and severe locomotor defects. Knockdown of RhoBTB in the Drosophila dendritic arborization neurons resulted in a decreased number of dendrites, thus suggesting a role of RhoBTB in dendritic development. We have established missense variants in the BTB-domain-encoding region of RHOBTB2 as causative for a developmental and epileptic encephalopathy and have elucidated the role of atypical Rho GTPase RhoBTB in Drosophila neurological function and possibly dendrite development.
Keywords: Drosophila melanogaster; RHOBTB2; epileptic Encephalopathy; intellectual disability; proteasom; ubiquitination.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. - PubMed
-
- Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. - PubMed
-
- Vissers L.E., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

