Strong Genomic and Phenotypic Heterogeneity in the Aeromonas sobria Species Complex
- PMID: 29276504
- PMCID: PMC5727048
- DOI: 10.3389/fmicb.2017.02434
Strong Genomic and Phenotypic Heterogeneity in the Aeromonas sobria Species Complex
Abstract
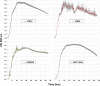
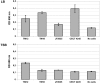
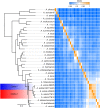
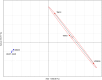
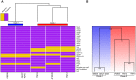
Aeromonas sobria is a mesophilic motile aeromonad currently depicted as an opportunistic pathogen, despite increasing evidence of mutualistic interactions in salmonid fish. However, the determinants of its host-microbe associations, either mutualistic or pathogenic, remain less understood than for other aeromonad species. On one side, there is an over-representation of pathogenic interactions in the A. sobria literature, of which only three articles to date report mutualistic interactions; on the other side, genomic characterization of this species is still fairly incomplete as only two draft genomes were published prior to the present work. Consequently, no study specifically investigated the biodiversity of A. sobria. In fact, the investigation of A. sobria as a species complex may have been clouded by: (i) confusion with A. veronii biovar sobria because of their similar biochemical profiles, and (ii) the intrinsic low resolution of previous studies based on 16S rRNA gene sequences and multilocus sequence typing. So far, the only high-resolution, phylogenomic studies of the genus Aeromonas included one A. sobria strain (CECT 4245 / Popoff 208), making it impossible to robustly conclude on the phylogenetic intra-species diversity and the positioning among other Aeromonas species. To further understand the biodiversity and the spectrum of host-microbe interactions in A. sobria as well as its potential genomic diversity, we assessed the genomic and phenotypic heterogeneity among five A. sobria strains: two clinical isolates recovered from infected fish (JF2635 and CECT 4245), one from an infected amphibian (08005) and two recently isolated brook charr probionts (TM12 and TM18) which inhibit in vitro growth of A. salmonicida subsp. salmonicida (a salmonid fish pathogen). A phylogenomic assessment including 2,154 softcore genes corresponding to 946,687 variable sites from 33 Aeromonas genomes confirms the status of A. sobria as a distinct species divided in two subclades, with 100% bootstrap support. The phylogenomic split of A. sobria in two subclades is corroborated by a deep dichotomy between all five A. sobria strains in terms of inhibitory effect against A. salmonicida subsp. salmonicida, gene contents and codon usage. Finally, the antagonistic effect of A. sobria strains TM12 and TM18 suggests novel control methods against A. salmonicida subsp. salmonicida.
Keywords: Aeromonas sobria; bacterial genomics; host–microbe interactions; microbial diversity; molecular systematics.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

