Genome sequence of the model plant pathogen Pectobacterium carotovorum SCC1
- PMID: 29276572
- PMCID: PMC5738896
- DOI: 10.1186/s40793-017-0301-z
Genome sequence of the model plant pathogen Pectobacterium carotovorum SCC1
Abstract
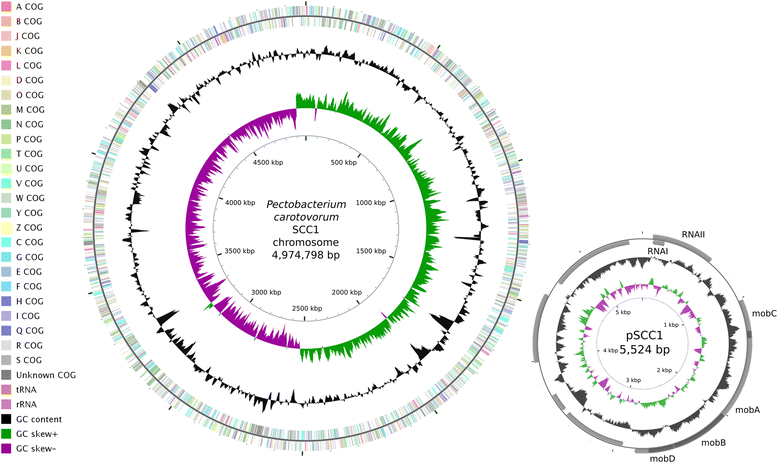
Bacteria of the genus Pectobacterium are economically important plant pathogens that cause soft rot disease on a wide variety of plant species. Here, we report the genome sequence of Pectobacterium carotovorum strain SCC1, a Finnish soft rot model strain isolated from a diseased potato tuber in the early 1980's. The genome of strain SCC1 consists of one circular chromosome of 4,974,798 bp and one circular plasmid of 5524 bp. In total 4451 genes were predicted, of which 4349 are protein coding and 102 are RNA genes.
Keywords: Finland; Necrotroph; Pectobacterium; Plant pathogen; Potato; Soft rot.
Conflict of interest statement
The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Pérombelon MCM. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 2002;51:1–12. doi: 10.1046/j.0032-0862.2001.Shorttitle.doc.x. - DOI
-
- Czajkowski R, Pérombelon MCM, van Veen JA, van der Wolf JM. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: a review. Plant Pathol. 2011;60:999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. - DOI
-
- Saarilahti HT, Palva ET. Major outer membrane proteins in the phytopathogenic bacteria Erwinia carotovora subsp. carotovora and subsp. atroseptica. FEMS Microbiol. Lett. 1986;35:267–270.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

