Directed evolution to improve protein folding in vivo
- PMID: 29278775
- PMCID: PMC5880552
- DOI: 10.1016/j.sbi.2017.12.003
Directed evolution to improve protein folding in vivo
Abstract
Recently, several innovative approaches have been developed that allow one to directly screen or select for improved protein folding in the cellular context. These methods have the potential of not just leading to a better understanding of the in vivo folding process, they may also allow for improved production of proteins of biotechnological interest.
Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
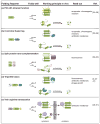
References
-
- DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. - PubMed
-
- Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JCA. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–871. Foit et al. developed a direct selection system for stable protein variants in vivo based on a tripartite fusion with a split antibiotic marker protein. Based on their isolated stabilized protein variants, it was concluded that thermodynamic and kinetic stability seem to be the decisive factors for expression and protein stability in vivo. - PMC - PubMed
-
- Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins Struct Funct Genet. 2002;46:105–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

