RNA Interference Therapies for an HIV-1 Functional Cure
- PMID: 29280961
- PMCID: PMC5795421
- DOI: 10.3390/v10010008
RNA Interference Therapies for an HIV-1 Functional Cure
Abstract
HIV-1 drug therapies can prevent disease progression but cannot eliminate HIV-1 viruses from an infected individual. While there is hope that elimination of HIV-1 can be achieved, several approaches to reach a functional cure (control of HIV-1 replication in the absence of drug therapy) are also under investigation. One of these approaches is the transplant of HIV-1 resistant cells expressing anti-HIV-1 RNAs, proteins or peptides. Small RNAs that use RNA interference pathways to target HIV-1 replication have emerged as competitive candidates for cell transplant therapy and have been included in all gene combinations that have so far entered clinical trials. Here, we review RNA interference pathways in mammalian cells and the design of therapeutic small RNAs that use these pathways to target pathogenic RNA sequences. Studies that have been performed to identify anti-HIV-1 RNA interference therapeutics are also reviewed and perspectives on their use in combination gene therapy to functionally cure HIV-1 infection are provided.
Keywords: HIV-1; RNA interference; cell transplant; functional cure; micro RNA; small/short hairpin RNA; small/short interfering RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
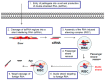
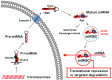
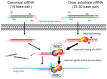
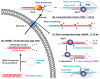
References
-
- Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R., et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

