Please do not recycle! Translation reinitiation in microbes and higher eukaryotes
- PMID: 29281028
- PMCID: PMC5972666
- DOI: 10.1093/femsre/fux059
Please do not recycle! Translation reinitiation in microbes and higher eukaryotes
Abstract
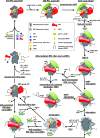
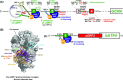
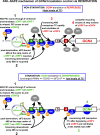
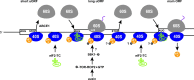
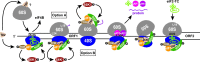
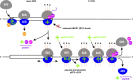
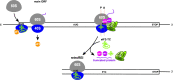
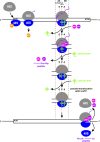
Protein production must be strictly controlled at its beginning and end to synthesize a polypeptide that faithfully copies genetic information carried in the encoding mRNA. In contrast to viruses and prokaryotes, the majority of mRNAs in eukaryotes contain only one coding sequence, resulting in production of a single protein. There are, however, many exceptional mRNAs that either carry short open reading frames upstream of the main coding sequence (uORFs) or even contain multiple long ORFs. A wide variety of mechanisms have evolved in microbes and higher eukaryotes to prevent recycling of some or all translational components upon termination of the first translated ORF in such mRNAs and thereby enable subsequent translation of the next uORF or downstream coding sequence. These specialized reinitiation mechanisms are often regulated to couple translation of the downstream ORF to various stimuli. Here we review all known instances of both short uORF-mediated and long ORF-mediated reinitiation and present our current understanding of the underlying molecular mechanisms of these intriguing modes of translational control.
Figures



References
-
- Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol 1990;213:811–8. - PubMed
-
- Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 2005;20:251–62. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

