The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ
- PMID: 29281621
- PMCID: PMC5760103
- DOI: 10.1371/journal.pgen.1007119
The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ
Erratum in
-
Correction: The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ.PLoS Genet. 2018 Feb 14;14(2):e1007236. doi: 10.1371/journal.pgen.1007236. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29444075 Free PMC article.
Abstract
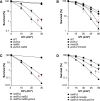
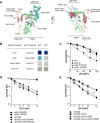
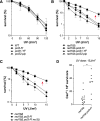
Replicative DNA polymerases cannot insert efficiently nucleotides at sites of base lesions. This function is taken over by specialized translesion DNA synthesis (TLS) polymerases to allow DNA replication completion in the presence of DNA damage. In eukaryotes, Rad6- and Rad18-mediated PCNA ubiquitination at lysine 164 promotes recruitment of TLS polymerases, allowing cells to efficiently cope with DNA damage. However, several studies showed that TLS polymerases can be recruited also in the absence of PCNA ubiquitination. We hypothesized that the stability of the interactions between DNA polymerase δ (Pol δ) subunits and/or between Pol δ and PCNA at the primer/template junction is a crucial factor to determine the requirement of PCNA ubiquitination. To test this hypothesis, we used a structural mutant of Pol δ in which the interaction between Pol3 and Pol31 is inhibited. We found that in yeast, rad18Δ-associated UV hypersensitivity is suppressed by pol3-ct, a mutant allele of the POL3 gene that encodes the catalytic subunit of replicative Pol δ. pol3-ct suppressor effect was specifically dependent on the Rev1 and Pol ζ TLS polymerases. This result strongly suggests that TLS polymerases could rely much less on PCNA ubiquitination when Pol δ interaction with PCNA is partially compromised by mutations. In agreement with this model, we found that the pol3-FI allele suppressed rad18Δ-associated UV sensitivity as observed for pol3-ct. This POL3 allele carries mutations within a putative PCNA Interacting Peptide (PIP) motif. We then provided molecular and genetic evidence that this motif could contribute to Pol δ-PCNA interaction indirectly, although it is not a bona fide PIP. Overall, our results suggest that the primary role of PCNA ubiquitination is to allow TLS polymerases to outcompete Pol δ for PCNA access upon DNA damage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae.DNA Repair (Amst). 2014 Dec;24:138-149. doi: 10.1016/j.dnarep.2014.04.013. Epub 2014 May 10. DNA Repair (Amst). 2014. PMID: 24819597 Free PMC article.
-
PCNA trimer instability inhibits translesion synthesis by DNA polymerase η and by DNA polymerase δ.DNA Repair (Amst). 2013 May 1;12(5):367-76. doi: 10.1016/j.dnarep.2013.02.007. Epub 2013 Mar 15. DNA Repair (Amst). 2013. PMID: 23506842 Free PMC article.
-
NMR mapping of PCNA interaction with translesion synthesis DNA polymerase Rev1 mediated by Rev1-BRCT domain.J Mol Biol. 2013 Sep 9;425(17):3091-105. doi: 10.1016/j.jmb.2013.05.029. Epub 2013 Jun 7. J Mol Biol. 2013. PMID: 23747975
-
Ubiquitin-dependent regulation of translesion polymerases.Biochem Soc Trans. 2010 Feb;38(Pt 1):110-5. doi: 10.1042/BST0380110. Biochem Soc Trans. 2010. PMID: 20074045 Review.
-
Polymerase Delta in Eukaryotes: How is It Transiently Exchanged with Specialized DNA Polymerases During Translesion DNA Synthesis?Curr Protein Pept Sci. 2018;19(8):790-804. doi: 10.2174/1389203719666180430155625. Curr Protein Pept Sci. 2018. PMID: 29708067 Review.
Cited by
-
Differential mechanisms of tolerance to extreme environmental conditions in tardigrades.Sci Rep. 2019 Oct 17;9(1):14938. doi: 10.1038/s41598-019-51471-8. Sci Rep. 2019. PMID: 31624306 Free PMC article.
-
Telomeric replication stress: the beginning and the end for alternative lengthening of telomeres cancers.Open Biol. 2022 Mar;12(3):220011. doi: 10.1098/rsob.220011. Epub 2022 Mar 9. Open Biol. 2022. PMID: 35259951 Free PMC article. Review.
-
Rad51 filaments assembled in the absence of the complex formed by the Rad51 paralogs Rad55 and Rad57 are outcompeted by translesion DNA polymerases on UV-induced ssDNA gaps.PLoS Genet. 2023 Feb 7;19(2):e1010639. doi: 10.1371/journal.pgen.1010639. eCollection 2023 Feb. PLoS Genet. 2023. PMID: 36749784 Free PMC article.
-
The inner side of yeast PCNA contributes to genome stability by mediating interactions with Rad18 and the replicative DNA polymerase δ.Sci Rep. 2022 Mar 25;12(1):5163. doi: 10.1038/s41598-022-09208-7. Sci Rep. 2022. PMID: 35338218 Free PMC article.
-
Nucleotide Excision Repair, XPA-1, and the Translesion Synthesis Complex, POLZ-1 and REV-1, Are Critical for Interstrand Cross-Link Repair in Caenorhabditis elegans Germ Cells.Biochemistry. 2020 Sep 29;59(38):3554-3561. doi: 10.1021/acs.biochem.0c00719. Epub 2020 Sep 18. Biochemistry. 2020. PMID: 32945661 Free PMC article.
References
-
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff R V, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73: 134–154. doi: 10.1128/MMBR.00034-08 - DOI - PMC - PubMed
-
- Pagès V, Fuchs RPP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21: 8957–8966. doi: 10.1038/sj.onc.1206006 - DOI - PubMed
-
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst). 2007;6: 891–899. doi: 10.1016/j.dnarep.2007.02.003 - DOI - PubMed
-
- Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419: 135–141. doi: 10.1038/nature00991 - DOI - PubMed
-
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458: 461–467. doi: 10.1038/nature07963 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

