Overexpression of Map3k7 activates sinoatrial node-like differentiation in mouse ES-derived cardiomyocytes
- PMID: 29281682
- PMCID: PMC5744947
- DOI: 10.1371/journal.pone.0189818
Overexpression of Map3k7 activates sinoatrial node-like differentiation in mouse ES-derived cardiomyocytes
Abstract
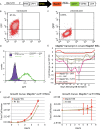
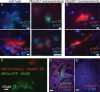
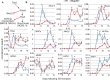
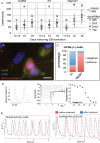
In vivo, cardiomyocytes comprise a heterogeneous population of contractile cells defined by unique electrophysiologies, molecular markers and morphologies. The mechanisms directing myocardial cells to specific sub-lineages remain poorly understood. Here we report that overexpression of TGFβ-Activated Kinase (TAK1/Map3k7) in mouse embryonic stem (ES) cells faithfully directs myocardial differentiation of embryoid body (EB)-derived cardiac cells toward the sinoatrial node (SAN) lineage. Most cardiac cells in Map3k7-overexpressing EBs adopt markers, cellular morphologies, and electrophysiological behaviors characteristic of the SAN. These data, in addition to the fact that Map3k7 is upregulated in the sinus venous-the source of cells for the SAN-suggest that Map3k7 may be an endogenous regulator of the SAN fate.
Conflict of interest statement
Figures
References
-
- Van Mierop LH, Gessner IH. The morphologic development of the sinoatrial node in the mouse. Am J Cardiol. 1970;25(2):204–12. Epub 1970/02/01. doi: 0002-9149(70)90580-1 [pii]. . - PubMed
-
- Barbuti A, Robinson RB. Stem cell-derived nodal-like cardiomyocytes as a novel pharmacologic tool: insights from sinoatrial node development and function. Pharmacol Rev. 2015;67(2):368–88. doi: 10.1124/pr.114.009597 . - DOI - PubMed
-
- Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, et al. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell. 1998;93(5):717–29. Epub 1998/06/18. doi: S0092-8674(00)81434-8 [pii]. . - PubMed
-
- Seifert R, Scholten A, Gauss R, Mincheva A, Lichter P, Kaupp UB. Molecular characterization of a slowly gating human hyperpolarization-activated channel predominantly expressed in thalamus, heart, and testis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):9391–6. Epub 1999/08/04. ; PubMed Central PMCID: PMC17793. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

