Global transcriptional landscape and promoter mapping of the gut commensal Bifidobacterium breve UCC2003
- PMID: 29281966
- PMCID: PMC5746004
- DOI: 10.1186/s12864-017-4387-x
Global transcriptional landscape and promoter mapping of the gut commensal Bifidobacterium breve UCC2003
Abstract
Background: Bifidobacterium breve represents a common member of the infant gut microbiota and its presence in the gut has been associated with host well being. For this reason it is relevant to investigate and understand the molecular mechanisms underlying the establishment, persistence and activities of this gut commensal in the host environment.
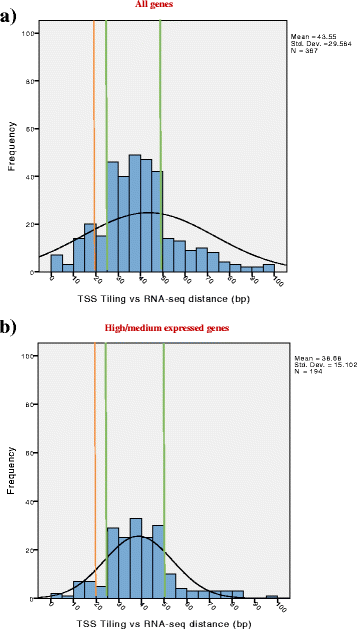
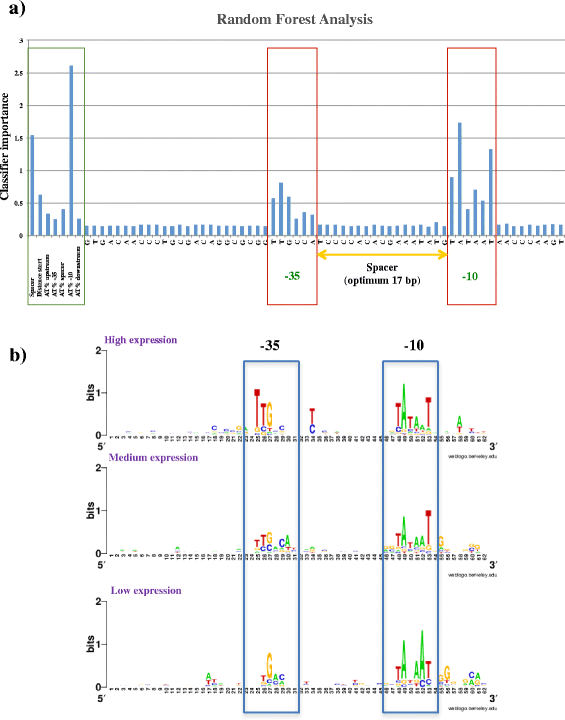
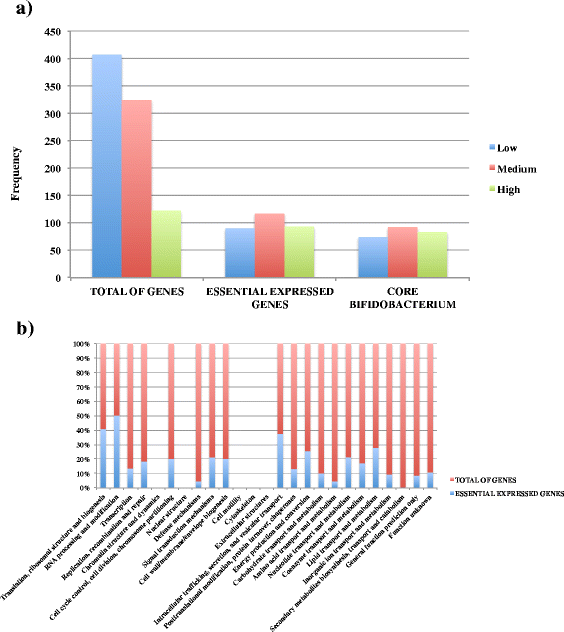
Results: The assessment of vegetative promoters in the bifidobacterial prototype Bifidobacterium breve UCC2003 was performed employing a combination of RNA tiling array analysis and cDNA sequencing. Canonical -10 (TATAAT) and -35 (TTGACA) sequences were identified upstream of transcribed genes or operons, where deviations from this consensus correspond to transcription level variations. A Random Forest analysis assigned the -10 region of B. breve promoters as the element most impacting on the level of transcription, followed by the spacer length and the 5'-UTR length of transcripts. Furthermore, our transcriptome study also identified rho-independent termination as the most common and effective termination signal of highly and moderately transcribed operons in B. breve.
Conclusion: The present study allowed us to identify genes and operons that are actively transcribed in this organism during logarithmic growth, and link promoter elements with levels of transcription of essential genes in this organism. As homologs of many of our identified genes are present across the whole genus Bifidobacterium, our dataset constitutes a transcriptomic reference to be used for future investigations of gene expression in members of this genus.
Keywords: Bifidobacteria; Gene expression; Probiotics; Transcription.
Conflict of interest statement
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Glycosulfatase-Encoding Gene Cluster in Bifidobacterium breve UCC2003.Appl Environ Microbiol. 2016 Oct 27;82(22):6611-6623. doi: 10.1128/AEM.02022-16. Print 2016 Nov 15. Appl Environ Microbiol. 2016. PMID: 27590817 Free PMC article.
-
Carbohydrate Syntrophy enhances the establishment of Bifidobacterium breve UCC2003 in the neonatal gut.Sci Rep. 2018 Jul 13;8(1):10627. doi: 10.1038/s41598-018-29034-0. Sci Rep. 2018. PMID: 30006512 Free PMC article.
-
Transcriptional landscape of the pMP7017 megaplasmid and its impact on the Bifidobacterium breve UCC2003 transcriptome.Microb Biotechnol. 2024 Jan;17(1):e14405. doi: 10.1111/1751-7915.14405. Epub 2024 Jan 11. Microb Biotechnol. 2024. PMID: 38206097 Free PMC article.
-
Bifidobacterium breve UCC2003 Employs Multiple Transcriptional Regulators To Control Metabolism of Particular Human Milk Oligosaccharides.Appl Environ Microbiol. 2018 Apr 16;84(9):e02774-17. doi: 10.1128/AEM.02774-17. Print 2018 May 1. Appl Environ Microbiol. 2018. PMID: 29500268 Free PMC article.
-
Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection.Gut Microbes. 2012 Sep-Oct;3(5):420-5. doi: 10.4161/gmic.20630. Epub 2012 Jun 20. Gut Microbes. 2012. PMID: 22713271 Review.
Cited by
-
Dissecting the Evolutionary Development of the Species Bifidobacterium animalis through Comparative Genomics Analyses.Appl Environ Microbiol. 2019 Mar 22;85(7):e02806-18. doi: 10.1128/AEM.02806-18. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30709821 Free PMC article.
-
Application of Recombinase-Based In Vivo Expression Technology to Bifidobacterium longum subsp. longum for Identification of Genes Induced in the Gastrointestinal Tract of Mice.Microorganisms. 2020 Mar 13;8(3):410. doi: 10.3390/microorganisms8030410. Microorganisms. 2020. PMID: 32183191 Free PMC article.
-
Molecular analysis of the replication functions of the bifidobacterial conjugative megaplasmid pMP7017.Microb Biotechnol. 2021 Jul;14(4):1494-1511. doi: 10.1111/1751-7915.13810. Epub 2021 May 3. Microb Biotechnol. 2021. PMID: 33939264 Free PMC article.
-
Gene Networks Underlying the Resistance of Bifidobacterium longum to Inflammatory Factors.Front Immunol. 2020 Nov 16;11:595877. doi: 10.3389/fimmu.2020.595877. eCollection 2020. Front Immunol. 2020. PMID: 33304352 Free PMC article.
-
Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut.Nat Microbiol. 2021 Nov;6(11):1367-1382. doi: 10.1038/s41564-021-00970-4. Epub 2021 Oct 21. Nat Microbiol. 2021. PMID: 34675385 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

