Polyamines in the life of Arabidopsis: profiling the expression of S-adenosylmethionine decarboxylase (SAMDC) gene family during its life cycle
- PMID: 29281982
- PMCID: PMC5745906
- DOI: 10.1186/s12870-017-1208-y
Polyamines in the life of Arabidopsis: profiling the expression of S-adenosylmethionine decarboxylase (SAMDC) gene family during its life cycle
Abstract
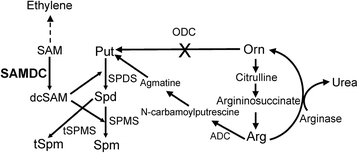
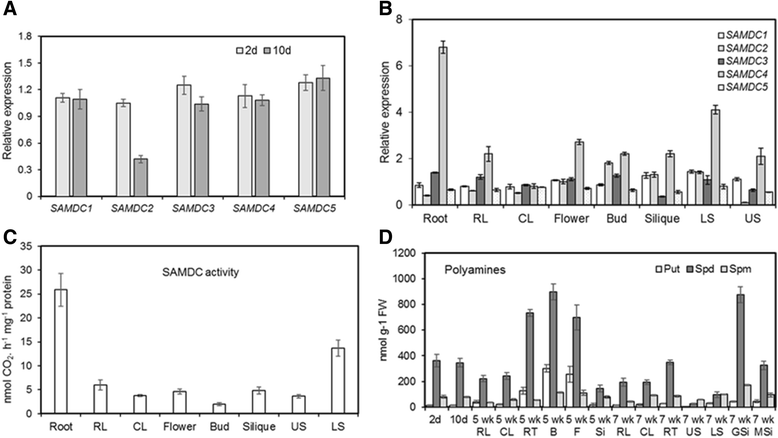
Background: Arabidopsis has 5 paralogs of the S-adenosylmethionine decarboxylase (SAMDC) gene. Neither their specific role in development nor the role of positive/purifying selection in genetic divergence of this gene family is known. While some data are available on organ-specific expression of AtSAMDC1, AtSAMDC2, AtSAMDC3 and AtSAMDC4, not much is known about their promoters including AtSAMDC5, which is believed to be non-functional.
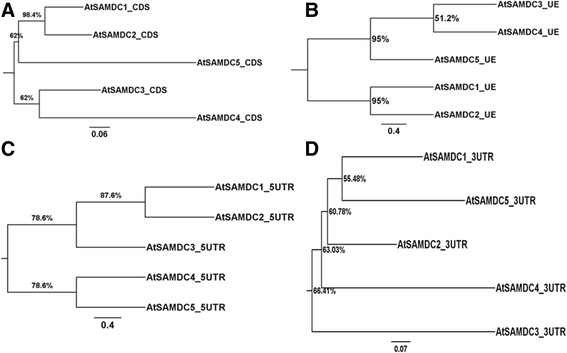
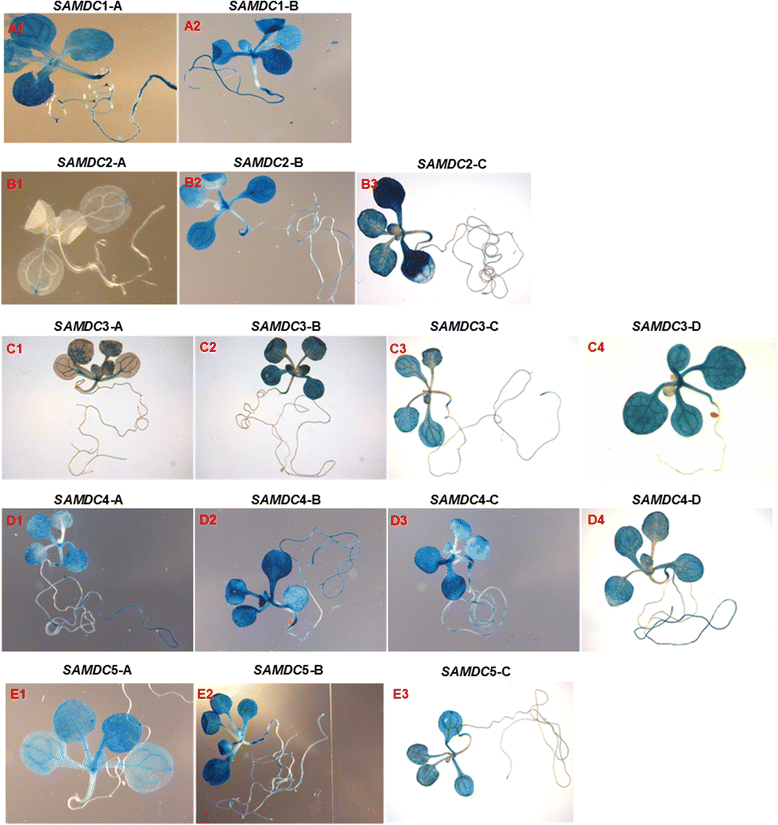
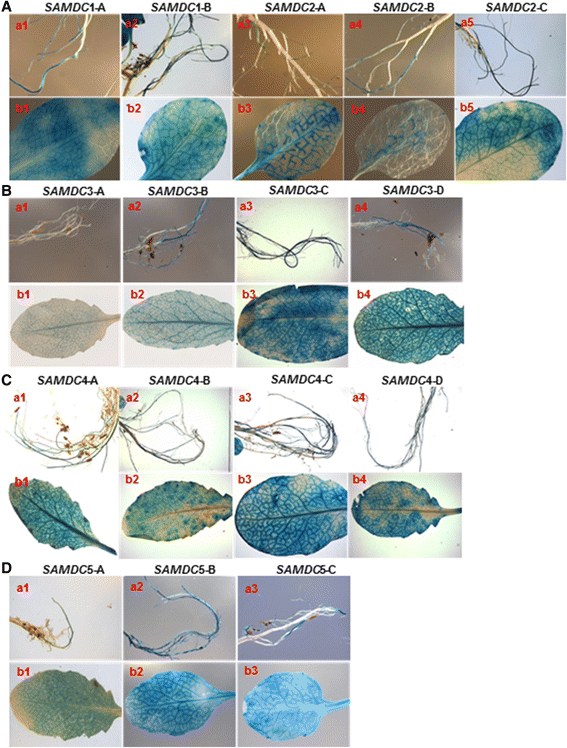
Results: (1) Phylogenetic analysis of the five AtSAMDC genes shows similar divergence pattern for promoters and coding sequences (CDSs), whereas, genetic divergence of 5'UTRs and 3'UTRs was independent of the promoters and CDSs; (2) while AtSAMDC1 and AtSAMDC4 promoters exhibit high activity (constitutive in the former), promoter activities of AtSAMDC2, AtSAMDC3 and AtSAMDC5 are moderate to low in seedlings (depending upon translational or transcriptional fusions), and are localized mainly in the vascular tissues and reproductive organs in mature plants; (3) based on promoter activity, it appears that AtSAMDC5 is both transcriptionally and translationally active, but based on it's coding sequence it seems to produce a non-functional protein; (4) though 5'-UTR based regulation of AtSAMDC expression through upstream open reading frames (uORFs) in the 5'UTR is well known, no such uORFs are present in AtSAMDC4 and AtSAMDC5; (5) the promoter regions of all five AtSAMDC genes contain common stress-responsive elements and hormone-responsive elements; (6) at the organ level, the activity of AtSAMDC enzyme does not correlate with the expression of specific AtSAMDC genes or with the contents of spermidine and spermine.
Conclusions: Differential roles of positive/purifying selection were observed in genetic divergence of the AtSAMDC gene family. All tissues express one or more AtSAMDC gene with significant redundancy, and concurrently, there is cell/tissue-specificity of gene expression, particularly in mature organs. This study provides valuable information about AtSAMDC promoters, which could be useful in future manipulation of crop plants for nutritive purposes, stress tolerance or bioenergy needs. The AtSAMDC1 core promoter might serve the need of a strong constitutive promoter, and its high expression in the gametophytic cells could be exploited, where strong male/female gametophyte-specific expression is desired; e.g. in transgenic modification of crop varieties.
Keywords: 3’UTR; 5’UTR; Arabidopsis; Gene family evolution; Metabolism; Polyamines; Promoter; SAMDC.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Franceschetti M, Hanfrey C, Scaramagli S, Torrigiani P, Bagni N, Burtin D, Michael AJ. Characterization of monocot and dicot plant S-adenosyl-l-methionine decarboxylase gene families including identification in the mRNA of a highly conserved pair of upstream overlapping open reading frames. Biochem J. 2001;353:403–409. doi: 10.1042/bj3530403. - DOI - PMC - PubMed
-
- WW H, Gong H, Pua EC. Molecular cloning and characterization of S-adenosyl-methionine decarboxylase genes from mustard (Brassica juncea) Physiol Plant. 2005;124:25–40. doi: 10.1111/j.1399-3054.2005.00500.x. - DOI
-
- Hao YJ, Zhang Z, Kitashiba H, Honda C, Ubi B, Kita M, Moriguchi T. Molecular cloning and functional characterization of two apple S-adenosylmethionine decarboxylase genes and their different expression in fruit development, cell growth and stress responses. Gene. 2005;350:41–50. doi: 10.1016/j.gene.2005.01.004. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

