Obesity alters the long-term fitness of the hematopoietic stem cell compartment through modulation of Gfi1 expression
- PMID: 29282250
- PMCID: PMC5789409
- DOI: 10.1084/jem.20170690
Obesity alters the long-term fitness of the hematopoietic stem cell compartment through modulation of Gfi1 expression
Abstract
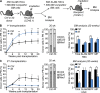
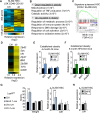
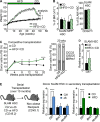
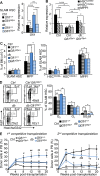
Obesity is a chronic organismal stress that disrupts multiple systemic and tissue-specific functions. In this study, we describe the impact of obesity on the activity of the hematopoietic stem cell (HSC) compartment. We show that obesity alters the composition of the HSC compartment and its activity in response to hematopoietic stress. The impact of obesity on HSC function is progressively acquired but persists after weight loss or transplantation into a normal environment. Mechanistically, we establish that the oxidative stress induced by obesity dysregulates the expression of the transcription factor Gfi1 and that increased Gfi1 expression is required for the abnormal HSC function induced by obesity. These results demonstrate that obesity produces durable changes in HSC function and phenotype and that elevation of Gfi1 expression in response to the oxidative environment is a key driver of the altered HSC properties observed in obesity. Altogether, these data provide phenotypic and mechanistic insight into durable hematopoietic dysregulations resulting from obesity.
© 2018 Lee et al.
Figures



References
-
- Ambrosi T.H., Scialdone A., Graja A., Gohlke S., Jank A.M., Bocian C., Woelk L., Fan H., Logan D.W., Schürmann A., et al. . 2017. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell. 20:771–784. 10.1016/j.stem.2017.02.009 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

