Arabidopsis thaliana rapid alkalinization factor 1-mediated root growth inhibition is dependent on calmodulin-like protein 38
- PMID: 29282286
- PMCID: PMC5808775
- DOI: 10.1074/jbc.M117.808881
Arabidopsis thaliana rapid alkalinization factor 1-mediated root growth inhibition is dependent on calmodulin-like protein 38
Abstract
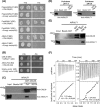
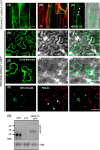
Arabidopsis thaliana rapid alkalinization factor 1 (AtRALF1) is a small secreted peptide hormone that inhibits root growth by repressing cell expansion. Although it is known that AtRALF1 binds the plasma membrane receptor FERONIA and conveys its signals via phosphorylation, the AtRALF1 signaling pathway is largely unknown. Here, using a yeast two-hybrid system to search for AtRALF1-interacting proteins in Arabidopsis, we identified calmodulin-like protein 38 (CML38) as an AtRALF1-interacting partner. We also found that CML38 and AtRALF1 are both secreted proteins that physically interact in a Ca2+- and pH-dependent manner. CML38-knockout mutants generated via T-DNA insertion were insensitive to AtRALF1, and simultaneous treatment with both AtRALF1 and CML38 proteins restored sensitivity in these mutants. Hybrid plants lacking CML38 and having high accumulation of the AtRALF1 peptide did not exhibit the characteristic short-root phenotype caused by AtRALF1 overexpression. Although CML38 was essential for AtRALF1-mediated root inhibition, it appeared not to have an effect on the AtRALF1-induced alkalinization response. Moreover, acridinium-labeling of AtRALF1 indicated that the binding of AtRALF1 to intact roots is CML38-dependent. In summary, we describe a new component of the AtRALF1 response pathway. The new component is a calmodulin-like protein that binds AtRALF1, is essential for root growth inhibition, and has no role in AtRALF1 alkalinization.
Keywords: calmodulin (CaM); cell growth; cell signaling; development; peptide hormone; rapid alkalinization factor; root; secreted protein; secretory pathway.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Mingossi F. B., Matos J. L., Rizzato A. P., Medeiros A. H., Falco M. C., Silva-Filho M. C., and Moura D. S. (2010) SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol. Biol. 73, 271–281 10.1007/s11103-010-9613-8 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

