Mechanism of N2 Reduction Catalyzed by Fe-Nitrogenase Involves Reductive Elimination of H2
- PMID: 29283553
- PMCID: PMC5837051
- DOI: 10.1021/acs.biochem.7b01142
Mechanism of N2 Reduction Catalyzed by Fe-Nitrogenase Involves Reductive Elimination of H2
Abstract
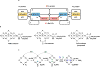
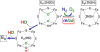
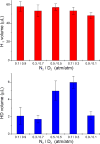
Of the three forms of nitrogenase (Mo-nitrogenase, V-nitrogenase, and Fe-nitrogenase), Fe-nitrogenase has the poorest ratio of N2 reduction relative to H2 evolution. Recent work on the Mo-nitrogenase has revealed that reductive elimination of two bridging Fe-H-Fe hydrides on the active site FeMo-cofactor to yield H2 is a key feature in the N2 reduction mechanism. The N2 reduction mechanism for the Fe-nitrogenase active site FeFe-cofactor was unknown. Here, we have purified both component proteins of the Fe-nitrogenase system, the electron-delivery Fe protein (AnfH) plus the catalytic FeFe protein (AnfDGK), and established its mechanism of N2 reduction. Inductively coupled plasma optical emission spectroscopy and mass spectrometry show that the FeFe protein component does not contain significant amounts of Mo or V, thus ruling out a requirement of these metals for N2 reduction. The fully functioning Fe-nitrogenase system was found to have specific activities for N2 reduction (1 atm) of 181 ± 5 nmol NH3 min-1 mg-1 FeFe protein, for proton reduction (in the absence of N2) of 1085 ± 41 nmol H2 min-1 mg-1 FeFe protein, and for acetylene reduction (0.3 atm) of 306 ± 3 nmol C2H4 min-1 mg-1 FeFe protein. Under turnover conditions, N2 reduction is inhibited by H2 and the enzyme catalyzes the formation of HD when presented with N2 and D2. These observations are explained by the accumulation of four reducing equivalents as two metal-bound hydrides and two protons at the FeFe-cofactor, with activation for N2 reduction occurring by reductive elimination of H2.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
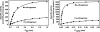


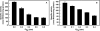
References
-
- Raymond J, Siefert JL, Staples CR, Blankenship RE. The natural history of nitrogen fixation. Mol. Biol. Evol. 2004;21:541–554. - PubMed
-
- Burris RH, Roberts GP. Biological nitrogen fixation. Annu. Rev. Nutr. 1993;13:317–35. - PubMed
-
- Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. - PubMed
-
- Eady RR. Structure-function relationships of alternative nitrogenases. Chem. Rev. 1996;96:3013–3030. - PubMed
-
- Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem. Rev. 1996;96:2983–3012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

