Retinol dehydrogenase-10 promotes development and progression of human glioma via the TWEAK-NF-κB axis
- PMID: 29285249
- PMCID: PMC5739636
- DOI: 10.18632/oncotarget.22166
Retinol dehydrogenase-10 promotes development and progression of human glioma via the TWEAK-NF-κB axis
Abstract
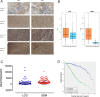
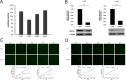
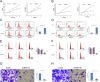
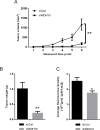
Retinol dehydrogenase-10 (RDH10) is a member of the short-chain dehydrogenase/reductase family, which plays an important role in retinoic acid (RA) synthesis. Here, we show that RDH10 is highly expressed in human gliomas, and its expression correlates with tumor grade and patient survival times. In vitro, lentivirus-mediated shRNA knockdown of RDH10 suppressed glioma cell proliferation, survival, and invasiveness and cell cycle progression. In vivo, RDH10 knockdown reduced glioma growth in nude mice. Microarray analysis revealed that RDH10 silencing reduces expression of TNFRSF12A (Fn14), TNFSF12 (TWEAK), TRAF3, IKBKB (IKK-β), and BMPR2, while it increases expression of TRAF1, NFKBIA (IκBα), NFKBIE (IκBε), and TNFAIP3. This suggests that RDH10 promotes glioma cell proliferation and survival by regulating the TWEAK-NF-κB axis, and that it could potentially serve as a novel target for human glioma treatment.
Keywords: glioma; nuclear factor kapaB (NF-κB); retinol dehydrogenase 10 (RDH10); the cancer genome atlas (TCGA); tumor necrosis factor-like weak inducer of apoptosis (TWEAK).
Conflict of interest statement
CONFLICTS OF INTEREST The authors have no conflicts of interest to disclose.
Figures
Similar articles
-
Retinol dehydrogenase 10 promotes metastasis of glioma cells via the transforming growth factor-β/SMAD signaling pathway.Chin Med J (Engl). 2019 Oct 20;132(20):2430-2437. doi: 10.1097/CM9.0000000000000478. Chin Med J (Engl). 2019. PMID: 31613821 Free PMC article.
-
Tumor necrosis factor-like weak inducer of apoptosis (TWEAK) promotes glioma cell invasion through induction of NF-κB-inducing kinase (NIK) and noncanonical NF-κB signaling.Mol Cancer. 2015 Jan 27;14(1):9. doi: 10.1186/s12943-014-0273-1. Mol Cancer. 2015. PMID: 25622756 Free PMC article.
-
Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome.Cancer Res. 2006 Oct 1;66(19):9535-42. doi: 10.1158/0008-5472.CAN-06-0418. Cancer Res. 2006. PMID: 17018610
-
FGF-inducible 14-kDa protein (Fn14) is regulated via the RhoA/ROCK kinase pathway in cardiomyocytes and mediates nuclear factor-kappaB activation by TWEAK.Basic Res Cardiol. 2010 Mar;105(2):301-13. doi: 10.1007/s00395-009-0046-y. Epub 2009 Jul 23. Basic Res Cardiol. 2010. PMID: 19629561
-
Role of tumor necrosis factor-like weak inducer of apoptosis (TWEAK)/fibroblast growth factor-inducible 14 (Fn14) axis in rheumatic diseases.Chin Med J (Engl). 2012 Nov;125(21):3898-904. Chin Med J (Engl). 2012. PMID: 23106895 Review.
Cited by
-
Ginkgo Flavonol Glycosides or Ginkgolides Tend to Differentially Protect Myocardial or Cerebral Ischemia-Reperfusion Injury via Regulation of TWEAK-Fn14 Signaling in Heart and Brain.Front Pharmacol. 2019 Jul 5;10:735. doi: 10.3389/fphar.2019.00735. eCollection 2019. Front Pharmacol. 2019. PMID: 31333457 Free PMC article.
-
Antioxidants and Therapeutic Targets in Ovarian Clear Cell Carcinoma.Antioxidants (Basel). 2021 Jan 28;10(2):187. doi: 10.3390/antiox10020187. Antioxidants (Basel). 2021. PMID: 33525614 Free PMC article. Review.
-
Retinol dehydrogenase 10 promotes epithelial-mesenchymal transition in spinal cord gliomas via PI3K/AKT pathway.Int J Immunopathol Pharmacol. 2024 Jan-Dec;38:3946320241276336. doi: 10.1177/03946320241276336. Int J Immunopathol Pharmacol. 2024. PMID: 39180753 Free PMC article.
-
Retinol dehydrogenase 10 promotes metastasis of glioma cells via the transforming growth factor-β/SMAD signaling pathway.Chin Med J (Engl). 2019 Oct 20;132(20):2430-2437. doi: 10.1097/CM9.0000000000000478. Chin Med J (Engl). 2019. PMID: 31613821 Free PMC article.
-
Identification of prognostic biomarkers related to retinoic acid metabolism in gliomas and analysis of their impact on the immune microenvironment.Medicine (Baltimore). 2024 Oct 11;103(41):e39836. doi: 10.1097/MD.0000000000039836. Medicine (Baltimore). 2024. PMID: 39465792 Free PMC article.
References
-
- Surawicz TS, McCarthy BJ, Kupelian V, Jukich PJ, Bruer JM, Davis FG. Descriptive epidemiology of primary brain and CNS tumors: results from the Central Brain Tumor Registry of the United States, 1990-1994. Neuro Oncol. 1999;1:14–25. https://doi.org/10.1093/neuonc/1.1.14 - DOI - PMC - PubMed
-
- Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncology. 2013(15):ii1–56. https://doi.org/10.1093/neuonc/not151 - DOI - PMC - PubMed
-
- Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613–621. https://doi.org/10.1016/j.cancergen.2012.10.009 - DOI - PubMed
-
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. https://doi.org/10.1001/jama.2013.280319 - DOI - PubMed
-
- Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. https://doi.org/10.1007/978-1-60327-492-0_14 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

