Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images
- PMID: 29285517
- PMCID: PMC5729919
- DOI: 10.1117/1.JMI.4.4.044504
Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images
Abstract
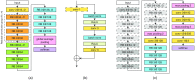
Currently, histopathological tissue examination by a pathologist represents the gold standard for breast lesion diagnostics. Automated classification of histopathological whole-slide images (WSIs) is challenging owing to the wide range of appearances of benign lesions and the visual similarity of ductal carcinoma in-situ (DCIS) to invasive lesions at the cellular level. Consequently, analysis of tissue at high resolutions with a large contextual area is necessary. We present context-aware stacked convolutional neural networks (CNN) for classification of breast WSIs into normal/benign, DCIS, and invasive ductal carcinoma (IDC). We first train a CNN using high pixel resolution to capture cellular level information. The feature responses generated by this model are then fed as input to a second CNN, stacked on top of the first. Training of this stacked architecture with large input patches enables learning of fine-grained (cellular) details and global tissue structures. Our system is trained and evaluated on a dataset containing 221 WSIs of hematoxylin and eosin stained breast tissue specimens. The system achieves an AUC of 0.962 for the binary classification of nonmalignant and malignant slides and obtains a three-class accuracy of 81.3% for classification of WSIs into normal/benign, DCIS, and IDC, demonstrating its potential for routine diagnostics.
Keywords: breast cancer; context-aware CNN; convolutional neural networks; deep learning; histopathology.
Figures



References
-
- Naik S., et al. , “Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology,” in 5th IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro (ISBI ’08), pp. 284–287 (2008).10.1109/ISBI.2008.4540988 - DOI
-
- Doyle S., et al. , “Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features,” in 5th IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro (ISBI ’08), pp. 496–499 (2008).10.1109/ISBI.2008.4541041 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

