Using single-cell multiple omics approaches to resolve tumor heterogeneity
- PMID: 29285690
- PMCID: PMC5746494
- DOI: 10.1186/s40169-017-0177-y
Using single-cell multiple omics approaches to resolve tumor heterogeneity
Abstract
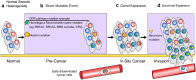
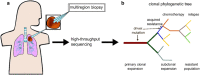
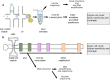
It has become increasingly clear that both normal and cancer tissues are composed of heterogeneous populations. Genetic variation can be attributed to the downstream effects of inherited mutations, environmental factors, or inaccurately resolved errors in transcription and replication. When lesions occur in regions that confer a proliferative advantage, it can support clonal expansion, subclonal variation, and neoplastic progression. In this manner, the complex heterogeneous microenvironment of a tumour promotes the likelihood of angiogenesis and metastasis. Recent advances in next-generation sequencing and computational biology have utilized single-cell applications to build deep profiles of individual cells that are otherwise masked in bulk profiling. In addition, the development of new techniques for combining single-cell multi-omic strategies is providing a more precise understanding of factors contributing to cellular identity, function, and growth. Continuing advancements in single-cell technology and computational deconvolution of data will be critical for reconstructing patient specific intra-tumour features and developing more personalized cancer treatments.
Keywords: Cancer; Gene expression; Heterogeneity; Methylation; Multi-omics; Mutation; Single-cell sequencing.
Figures

Similar articles
-
Advancements and applications of single-cell multi-omics techniques in cancer research: Unveiling heterogeneity and paving the way for precision therapeutics.Biochem Biophys Rep. 2023 Nov 29;37:101589. doi: 10.1016/j.bbrep.2023.101589. eCollection 2024 Mar. Biochem Biophys Rep. 2023. PMID: 38074997 Free PMC article. Review.
-
Multi-site tumor sampling highlights molecular intra-tumor heterogeneity in malignant pleural mesothelioma.Genome Med. 2021 Jul 14;13(1):113. doi: 10.1186/s13073-021-00931-w. Genome Med. 2021. PMID: 34261524 Free PMC article.
-
Cell subpopulation deconvolution reveals breast cancer heterogeneity based on DNA methylation signature.Brief Bioinform. 2017 May 1;18(3):426-440. doi: 10.1093/bib/bbw028. Brief Bioinform. 2017. PMID: 27016391
-
Intra-tumour heterogeneity - going beyond genetics.FEBS J. 2016 Jun;283(12):2245-58. doi: 10.1111/febs.13705. Epub 2016 Apr 1. FEBS J. 2016. PMID: 26945550 Review.
-
Advances in bulk and single-cell multi-omics approaches for systems biology and precision medicine.Brief Bioinform. 2021 Sep 2;22(5):bbab024. doi: 10.1093/bib/bbab024. Brief Bioinform. 2021. PMID: 33778867 Review.
Cited by
-
Construction of a Comprehensive Multiomics Map of Hepatocellular Carcinoma and Screening of Possible Driver Genes.Front Genet. 2020 Jun 25;11:634. doi: 10.3389/fgene.2020.00634. eCollection 2020. Front Genet. 2020. PMID: 32670354 Free PMC article.
-
Developmental origins and oncogenic pathways in malignant brain tumors.Wiley Interdiscip Rev Dev Biol. 2019 Jul;8(4):e342. doi: 10.1002/wdev.342. Epub 2019 Apr 3. Wiley Interdiscip Rev Dev Biol. 2019. PMID: 30945456 Free PMC article. Review.
-
Single cell transcriptome research in human placenta.Reproduction. 2020 Dec;160(6):R155-R167. doi: 10.1530/REP-20-0231. Reproduction. 2020. PMID: 33112783 Free PMC article. Review.
-
Machine learning approaches to drug response prediction: challenges and recent progress.NPJ Precis Oncol. 2020 Jun 15;4:19. doi: 10.1038/s41698-020-0122-1. eCollection 2020. NPJ Precis Oncol. 2020. PMID: 32566759 Free PMC article. Review.
-
Artificial-cell-type aware cell-type classification in CITE-seq.Bioinformatics. 2020 Jul 1;36(Suppl_1):i542-i550. doi: 10.1093/bioinformatics/btaa467. Bioinformatics. 2020. PMID: 32657383 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
