Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR
- PMID: 29286103
- PMCID: PMC5807040
- DOI: 10.3892/ijo.2017.4233
Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR
Abstract
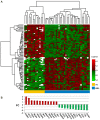
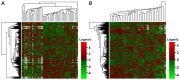
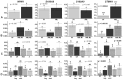
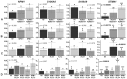
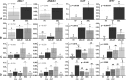
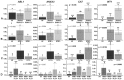
Acute myeloid leukemia (AML) is the most common and severe form of acute leukemia diagnosed in adults. Owing to its heterogeneity, AML is divided into classes associated with different treatment outcomes and specific gene expression profiles. Based on previous studies on AML, in this study, we designed and generated an AML-array containing 900 oligonucleotide probes complementary to human genes implicated in hematopoietic cell differentiation and maturation, proliferation, apoptosis and leukemic transformation. The AML-array was used to hybridize 118 samples from 33 patients with AML of the M1 and M2 subtypes of the French-American‑British (FAB) classification and 15 healthy volunteers (HV). Rigorous analysis of the microarray data revealed that 83 genes were differentially expressed between the patients with AML and the HV, including genes not yet discussed in the context of AML pathogenesis. The most overexpressed genes in AML were STMN1, KITLG, CDK6, MCM5, KRAS, CEBPA, MYC, ANGPT1, SRGN, RPLP0, ENO1 and SET, whereas the most underexpressed genes were IFITM1, LTB, FCN1, BIRC3, LYZ, ADD3, S100A9, FCER1G, PTRPE, CD74 and TMSB4X. The overexpression of the CPA3 gene was specific for AML with mutated NPM1 and FLT3. Although the microarray-based method was insufficient to differentiate between any other AML subgroups, quantitative PCR approaches enabled us to identify 3 genes (ANXA3, S100A9 and WT1) whose expression can be used to discriminate between the 2 studied AML FAB subtypes. The expression levels of the ANXA3 and S100A9 genes were increased, whereas those of WT1 were decreased in the AML-M2 compared to the AML-M1 group. We also examined the association between the STMN1, CAT and ABL1 genes, and the FLT3 and NPM1 mutation status. FLT3+/NPM1- AML was associated with the highest expression of STMN1, and ABL1 was upregulated in FLT3+ AML and CAT in FLT3- AML, irrespectively of the NPM1 mutation status. Moreover, our results indicated that CAT and WT1 gene expression levels correlated with the response to therapy. CAT expression was highest in patients who remained longer under complete remission, whereas WT1 expression increased with treatment resistance. On the whole, this study demonstrates that the AML-array can potentially serve as a first-line screening tool, and may be helpful for the diagnosis of AML, whereas the differentiation between AML subgroups can be more successfully performed with PCR-based analysis of a few marker genes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, et al. European LeukemiaNet Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. - DOI - PubMed
-
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. - DOI - PubMed
-
- Döhner H. Hematology Am Soc Hematol Educ Program. Washington, DC: 2007. Implication of the molecular characterization of acute myeloid leukemia; pp. 412–419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

