Signaling Pathways Driving Aberrant Splicing in Cancer Cells
- PMID: 29286307
- PMCID: PMC5793162
- DOI: 10.3390/genes9010009
Signaling Pathways Driving Aberrant Splicing in Cancer Cells
Abstract
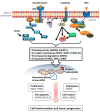
Aberrant profiles of pre-mRNA splicing are frequently observed in cancer. At the molecular level, an altered profile results from a complex interplay between chromatin modifications, the transcriptional elongation rate of RNA polymerase, and effective binding of the spliceosome to the generated transcripts. Key players in this interplay are regulatory splicing factors (SFs) that bind to gene-specific splice-regulatory sequence elements. Although mutations in genes of some SFs were described, a major driver of aberrant splicing profiles is oncogenic signal transduction pathways. Signaling can affect either the transcriptional expression levels of SFs or the post-translational modification of SF proteins, and both modulate the ratio of nuclear versus cytoplasmic SFs in a given cell. Here, we will review currently known mechanisms by which cancer cell signaling, including the mitogen-activated protein kinases (MAPK), phosphatidylinositol 3 (PI3)-kinase pathway (PI3K) and wingless (Wnt) pathways but also signals from the tumor microenvironment, modulate the activity or subcellular localization of the Ser/Arg rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) families of SFs.
Keywords: alternative splicing; cancer cell; genetic program; signaling pathway; tumor microenvironment; tumorigenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

