Molecular screening of tsetse flies and cattle reveal different Trypanosoma species including T. grayi and T. theileri in northern Cameroon
- PMID: 29287598
- PMCID: PMC5747950
- DOI: 10.1186/s13071-017-2540-7
Molecular screening of tsetse flies and cattle reveal different Trypanosoma species including T. grayi and T. theileri in northern Cameroon
Abstract
Background: African trypanosomes are mainly transmitted through the bite of tsetse flies (Glossina spp.). The present study investigated the occurrence of pathogenic trypanosomes in tsetse flies and cattle in tsetse fly-infested areas of Northern Cameroon.
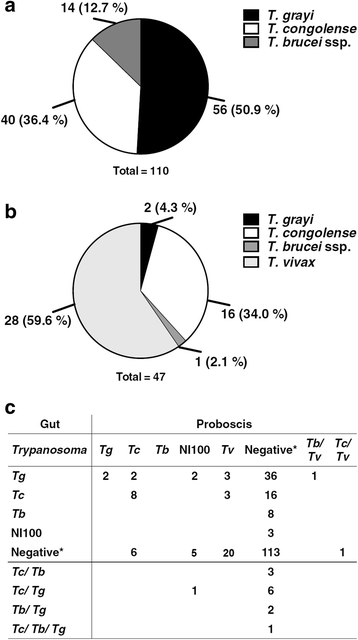
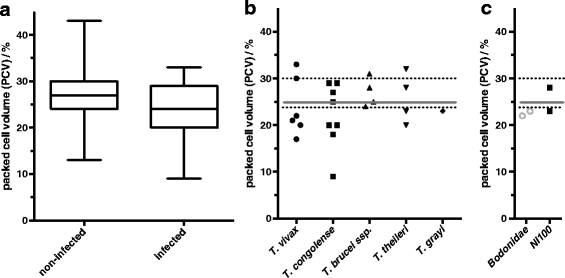
Results: Trypanosomes were identified using nested polymerase chain reaction (PCR) analysis of internal transcribed spacer 1 (ITS1) region, both by size estimation and sequencing of PCR products. Apparent density indices recorded in Gamba and Dodeo were 3.1 and 3.6 tsetse flies per trap and day, respectively. Trypanosoma prevalence infection rate for the tsetse fly gut (40%) and proboscis (19%) were recorded. Among the flies where trypanosomes were detected in the gut, 41.7% were positive for T. congolense and 14.6% for T. brucei ssp., whereas in the proboscis 36% harboured T. congolense and 62% contained T. vivax. T. grayi was highly prevalent in tsetse fly gut (58%). The most common mixed infections were the combination of T. congolense and T. grayi. Trypanosome prevalence rate in cattle blood was 6%. Among these, T. vivax represented 26%, T. congolense 35%, T. brucei ssp. 17% and T. theileri 17% of the infections. Surprisingly, in one case T. grayi was found in cattle. The mean packed cell volume (PCV) of cattle positive for trypanosomes was significantly lower (24.1 ± 5.6%; P < 0.05) than that of cattle in which trypanosomes were not detected (27.1 ± 4.9%). Interestingly, the occurrence of T. theileri or T. grayi DNA in cattle also correlated with low PCV at pathological levels.
Conclusion: This molecular epidemiological study of Trypanosoma species in Northern Cameroon revealed active foci of trypanosomes in Dodeo and Gamba. These findings are relevant in assessing the status of trypanosomosis in these regions and will serve as a guide for setting the priorities of the government in the control of the disease.
Keywords: Bodonidae; Cattle; ITS1; Northern Cameroon; Trypanosoma grayi; Trypanosoma theileri; Trypanosomosis; Tsetse fly.
Conflict of interest statement
Ethics approval
The study was conducted with the approval of the Ministère de l’élévage des pêches et des industries animales (MINEPIA) and by Mission spéciale d’éradication des glossines (MSEG) at the National and district levels, as well as the district veterinary officers in each of the study districts.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

